Vn8.4 GA4.0 Release Candidate: RC6.0
This page documents the vn8.4 GA4.0 release candidate RC6.0 (xkawa/f on MONSooN and xkawe on ARCHER). This job has been developed and run by Luke Abraham.
This job is NOT SUITABLE FOR RELEASE and SHOULD NOT BE USED FOR SCIENTIFIC PURPOSES. As well as the amber warnings given below for stratospheric chemistry and tropospheric and stratospheric aerosol, technical reasons (non-uniform polar rows) mean that this job should not be used.
Suitability for Release
| Tropospheric Chemistry | |
| Stratospheric Chemistry | Users should note the low stratospheric NOy. |
| Tropospheric Aerosol | Users should note the high aerosol optical depth in this configuration. |
| Stratospheric Aerosol | Users should note the high stratospheric sea-salt mixing ratios. |
Key
| Release candidate still under evaluation |
| Release candidate not scientifically suitable to be released |
| Release candidate is suitable for development jobs, and may be scientifically suitable to be released for some applications |
| Release candidate scientifically suitable to be released |
| Release candidate does not consider the required chemistry or aerosol processes |
Overview
MONSooN job xkawa(/f) |
ARCHER job xkawe
| |
| UKCA branch used | fcm:um_br/pkg/Config/vn8.4_UKCA@16485 (xkawf) |
fcm:um_br/pkg/Config/vn8.4_UKCA@16485
|
| Decomposition | 12EW x 16NS on 6 nodes (see below) | 12EW x 12NS on 12 nodes (see below) |
| Run-time (per model month) | ~2 hours (see below) | ~2 hours (see below) |
| Job-step (recommended) | 1 model month in 10800 seconds (3 hours) | 3 model months in 28800 seconds (8 hours) |
| Cost (per model year) | 150 node-hours (see below) | 115 kAU (see below) |
| Storage Requirements (current STASH settings) |
110GB per model year (32-bit pp-files & seasonal 64-bit dumps) MOOSE costs: £12.01 per model year |
110GB per model year (32-bit pp-files & seasonal 64-bit dumps) copied to the /nerc disk as the model runs,using the branch fcm:um_br/dev/luke/vn8.4_hector_monsoon_archiving_ff2pp/src as a central script modification.
|
UKCA code
For this release the relevant and specific UKCA code changes (when compared to the trunk) have been merged into a package branch on PUMA
For the MONSooN results presented here, revision number 16065 was used. The latest revision is 16485 (see below) and was used for the ARCHER results.
For details as to which branches and fixes were included, please see PUMA Trac ticket #632.
Changes since xkawa run
There have been two changes to this branch since job xkawa was completed. These have been tested by comparing a 1-month run of xkawa-identical job xkawg and running cumf on the 1st January dump, comparing with the equivalent job with these changes in, xkawf. These changes do not affect the model evolution.
- Deallocation of
qsmr: When porting this job to ARCHER the Cray cce Fortran compiler picked up that the variableqsmrwas deallocated too early. The IBM xlf Fortran compiler allows deallocated arrays still to be accessed, but the Cray compiler does not (correctly!). This bug was corrected at revision number r16244. - ARCHER specific changes: There were two changes to this congfiguration that were found by Karthee Sivalingam when he ported job
xjcimto ARCHER, which he placed in the branch fcm:um_br/dev/karthee/vn8.4_xjcim_port_fixes. I have merged this into the package branch at r16246. - Bugfixes from CSIRO: On porting this job to the CSIRO systems, two bugs were found. Peter Uhe of CSIRO has provided a patchfile which has been merged into the package branch at revision r16485. The bugs found were:
- Duplicated declaration of
i_mode_nucscavinsrc/atmosphere/UKCA/ukca_option_mod.F90. - Removal of windows line-breaks from
src/atmosphere/UKCA/ukca_radaer_read_precalc.F90.
- Duplicated declaration of
The cumf summary is below. While it says "files DO NOT compare" this is not due to any changes in the data fields, which are identical (as expected).
$ /projects/um1/vn8.4/ibm/utils/cumf xkawf/xkawfa.da20000101_00 xkawg/xkawga.da20000101_00 COMPARE - SUMMARY MODE ----------------------- Number of fields in file 1 = 47392 Number of fields in file 2 = 47392 Number of fields compared = 47392 FIXED LENGTH HEADER: Number of differences = 3 INTEGER HEADER: Number of differences = 0 REAL HEADER: Number of differences = 0 LEVEL DEPENDENT CONSTANTS: Number of differences = 0 LOOKUP: Number of differences = 33715 DATA FIELDS: Number of fields with differences = 0 files DO NOT compare
Using this branch
Note: if you wish to use this branch to develop some extra code, please follow these guidelines:
- Make your own branch (in the usual way) at vn8.4.
fcm mergein the UKCA package branchfcm:um_br/pkg/Config/vn8.4_UKCAat the latest revision.fcm committhis before you make your own changes.- In the UMUI, turn off the UKCA package branch, and use your own branch that you have just made.
- It is advisable to use a working copy initially while you are getting any developments working.
- For production runs it is best to
fcm commitall developments from the working copy and run from the repository using this revision number.- This will make it easier to repeat simulations at a later date if needed.
- Remember to perform frequent commits, even if you are still using the working copy - this makes it easier to backtrack changes when necessary.
Do not checkout the package branch, make changes, and commit them. This will cause problems for other UKCA users.
Functionality
Base Model
The base atmosphere model used here is the GA4.0 configuration. More information on GA4.0 development can be found on Global Atmosphere 4.0/Global Land 4.0 documentation pages (password required). A GMD paper documenting this model is also available.
The configuration is based on the Met Office job anenj (via MONSooN job xhmaj) which is derived from amche (the standard GA4.0 N96L85 interactive dust model) via
amche (base GA4.0 job) owned by Dan Copsey
akwxo (UKCA turned on) owned by Mohit Dalvi
aneni owned by Colin Johnson
anenj
xhmaj owned by Mohit Dalvi
xjcib (HOx recycling added) owned by Luke Abraham
xjcie (some reaction rates updated)
xjcih (O3 now interactive with radiation scheme)
xjcim (made TS2000 - see Initial conditions and forcing)
xjcin
xjlla (UKCA Tutorials base job; some branch consolidation) owned by Luke Abraham
xkawa (package branch used, containing various bugfixes and additions) owned by Luke Abraham
xkawf (further changes as described above)
For more information on these jobs and the UKCA release cycle, please see the the developing releases page.
Scaling (MONSooN)
Each compute node of the MONSooN phase 2 system contains four 3.8 GHz IBM Power 7 processors, and there is 64GB of RAM per node. Due to memory restrictions, UKCA is unable to run on less than 3 nodes. As noted below, the EW domain decomposition needed to be a multiple of 12. All simulations used 2 OpenMP threads which will not reduce the number of cores per node used.
More information on MONSooN can be found on the collaboration twiki (registration required).
12EW Decomposition
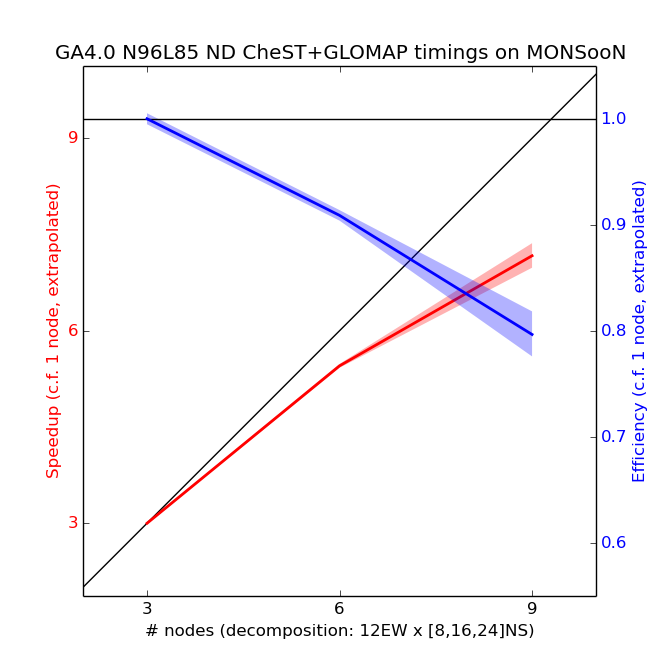
Scaling tests have been done from 3 to 9 nodes of MONSooN using a decomposition of 12EW, with a series of 1-day runs, with the results presented below.
The speedup in the plot above is calculated by assuming a linear scaling from 3 nodes down to 1 node. 5 simulations were performed for each number of nodes, and the envelope is 2 standard deviations (assuming that the standard deviation of the extrapolated 1-node data-point is the mean of the standard deviations of all other points). From these tests the recommended decomposition is 12 EW x 16 NS (i.e. 6 nodes, or 192 cores). Running on 6 nodes means that the model will complete 1 model month in approximately 2 hours. Although 3 nodes would be slightly more efficient, each simulation would take nearly twice as long and would therefore fall outside the 3-hour queue limit, which is undesirable. Also, MONSooN only has a maximum of 149 compute nodes available at any one time (4768 cores), and so larger jobs are likely to queue for longer. For this reason it is best to use the smallest number of nodes that still allows a job-step to run in 3 hours, while still being close to linear speedup.
For FairShare (login required) estimations, this job requires 150 node-hours per model year, accounting for slight variations in run-time.
Above 9 nodes the model not run as this is a 12EWx32NS which falls over due to the halo size in the model with the error message
???????????????????????????????????????????????????????????????????????????????? ???!!!???!!!???!!!???!!!???!!!???!!! ERROR ???!!!???!!!???!!!???!!!???!!!???!!!? ? Error in routine: DECOMP_DB:DECOMPOSE ? Error Code: 5 ? Error Message: Too many processors in the North-South direction ( 32) to support the extended halo size ( 5). Try running with 28 processors. ? Error generated from processor: 0 ? This run generated 5 warnings ????????????????????????????????????????????????????????????????????????????????
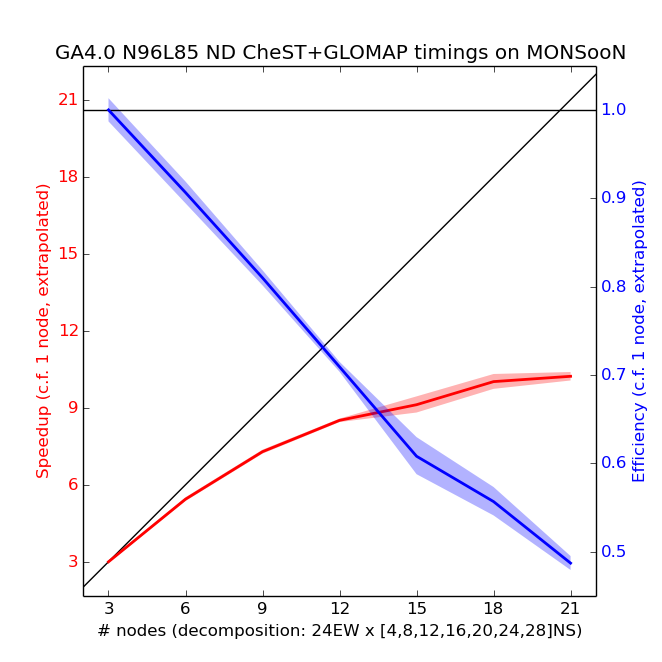
24EW Decomposition
Scaling tests have been done from 3 to 21 nodes of MONSooN using a decomposition of 24EW, with a series of 1-day runs, with the results presented below.
The speedup in the plot above is calculated by assuming a linear scaling from 3 nodes down to 1 node. 5 simulations were performed for each number of nodes, and the envelope is 2 standard deviations (assuming that the standard deviation of the extrapolated 1-node data-point is the mean of the standard deviations of all other points). Note the change in y scales when compared to the above plot for 12EW decomposition.
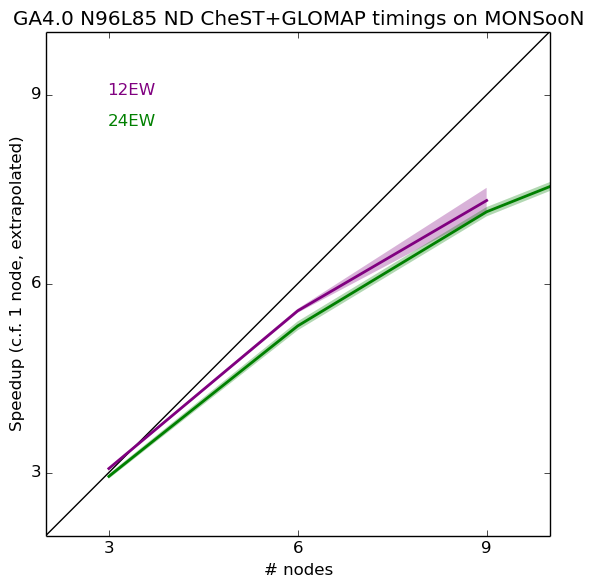
12EW verses 24EW decomposition
In the plots above, the extrapolated 1-node value is calculated from the 12EW or 24EW 3-node value. In reality, these are actually slightly different. If the 1-node value is calculated as a mean of the 12EW and 24EW values (with the standard deviation calculated accordingly) we can then see that using a 12EW decomposition is more advantageous over the 24EW decomposition (shadings are 2 standard deviations).
Therefore the decomposition that should be used is 12 EW x 16 NS, giving 1 model month in around 2 hours, and 150 node-hours per model year (for FairShare estimates).
Chemistry
This job uses the CheST chemistry scheme - an amalgamation of the Stratospheric Chemistry scheme and the Tropospheric Chemistry with Isoprene scheme. The Fast-JX interactive photolysis scheme is used.
For more information on these schemes, please see
- Evaluation of the new UKCA climate-composition model – Part 1: The stratosphere
- Evaluation of the new UKCA climate-composition model – Part 2: The Troposphere
- Implementation of the Fast-JX Photolysis scheme (v6.4) into the UKCA component of the MetUM chemistry-climate model (v7.3)
Aerosols
This job uses an extension to the GLOMAP-mode aerosol scheme which extends it into the stratosphere, although it is also suitable for tropospheric work.
For information on GLOMAP-mode, please see
- Description and evaluation of GLOMAP-mode: a modal global aerosol microphysics model for the UKCA composition-climate model
- Impact of the modal aerosol scheme GLOMAP-mode on aerosol forcing in the Hadley Centre Global Environmental Model
- Aerosol microphysics simulations of the Mt. Pinatubo eruption with the UKCA composition-climate model
Pressure-level output for tracers and chemical diagnostics
All STASH items available in section 34 have an equivalent in section 35. Currently only O3 and the flux through the CH4+OH reaction have been set-up, but other fields can be output by copying these examples.
Note: the Heaviside function s35i173 also need to put output so that these fields can be processed correctly to remove points which should contain missing data. In the lower levels of the model the pressures will be low (especially over high orography) and these points need to be masked off. Valid points are where this function is equal to 1.
RCP Scenario Code
The RCP scenario branch, committed to the trunk at vn8.6, has equivalent functionality available in this job (UKCA lower boundary conditions only - further changes are required to have these affect the GHGs).
To use this functionality a hand-edit is required - see e.g. /home/ukca/hand_edits/VN8.4/UKCA_RCP6.0.ed
Pre-compiled builds
This job makes use of pre-compiled builds, as can be seen in the FCM panel. This decreases the time required for the compilation step.
Note that the reconfiguration executable (qxreconf) has also been pre-compiled (this can be seen in the Compile and run options for the Atmosphere and Reconfiguration panel). This was required to allow the pre-compiled builds to work. If you want/need to change either the reconfiguration executable or the use of pre-compiled builds, you will need to change the settings in both the FCM and compilation options panels. Do not turn on compilation of the reconfiguration executable unless you change its location.
cumf tests (MONSooN)
A handy test of a model is to know whether it bit-compares when re-running, and what restrictions (if any) there are on this. To do this, the cumf utility is used, which is found at /projects/um1/vn8.4/ibm/utils/cumf. There are 4 types of tests that should be run, a NRUN-NRUN test, a NRUN-CRUN test, a CRUN-CRUN test, and a change of dump frequency test.
Note: A change of domain decomposition test was not performed as it is known that this configuration of UKCA will fail this test. This is due to the way that the Newton Raphson chemical solver converges, which is done over the whole domain. While the results would be different across different domain decompositions, they are all scientifically valid. So long as the decomposition is not changed during a run, results will be comparable (also taking the results of the other tests into account).
All tests were run without STASH, climate meaning, or the UKCA evaluation suite hand-edit (~mdalvi/umui_jobs/hand_edits/vn8.4/add_ukca_eval1_diags_l85.ed), as all of these can affect the dump by placing temporary fields in it. The model was run for 2 days, either with daily or 2-day dumping, and for the NRUN-CRUN test, the first step was for 1 day followed by a new job step for the 2nd day. Reconfiguration was only run once, at the start of the 1st test, and after that point the same .astart file was used by all jobs.
The following cumf tests were performed using revision r16246. It is not anticipated that the changes in r16485 will change these.
NRUN-NRUN tests (MONSooN)
For this test the model is run twice. The 2-day dumps are then compared. For this test it makes no difference if you compare the dumps produced using daily-dumping or 2-day dumping, as both pass this test (when compared to the equivalent dump produced using the same dumping frequency).
COMPARE - SUMMARY MODE ----------------------- Number of fields in file 1 = 14272 Number of fields in file 2 = 14272 Number of fields compared = 14272 FIXED LENGTH HEADER: Number of differences = 3 INTEGER HEADER: Number of differences = 0 REAL HEADER: Number of differences = 0 LEVEL DEPENDENT CONSTANTS: Number of differences = 0 LOOKUP: Number of differences = 0 DATA FIELDS: Number of fields with differences = 0 files compare, ignoring Fixed Length Header
NRUN-CRUN tests (MONSooN)
For this test the 2nd day dump of a daily dumping run (which has been run in a single jobstep) is compared with the 2nd day dump of a run where this was produced on a CRUN step (i.e. where the 1st day dump was produced on the NRUN step). In this case, the dumps DO NOT compare.
COMPARE - SUMMARY MODE ----------------------- Number of fields in file 1 = 14272 Number of fields in file 2 = 14272 Number of fields compared = 14272 FIXED LENGTH HEADER: Number of differences = 4 INTEGER HEADER: Number of differences = 0 REAL HEADER: Number of differences = 3 LEVEL DEPENDENT CONSTANTS: Number of differences = 0 LOOKUP: Number of differences = 0 DATA FIELDS: Number of fields with differences = 12221 Field 1 : Stash Code 2 : U COMPNT OF WIND AFTER TIMESTEP : Number of differences = 27840 ... ... Field 14076 : Stash Code 38405 : DRY PARTICLE DIAMETER AITKEN-INS : Number of differences = 67 files DO NOT compare
This is not unexpected for UKCA, as there are many variables which are initialised at the start of a run, but not saved to a dump to be re-initialised correctly. This is a feature which will need to be addressed in the future.
It should be noted that this does not invalidate a run, or prevent you from re-running to fill-in data gaps. You should, however, ensure that you maintain the original job-step length. On MONSooN is it recommended that maintain a 1-month jobstep length, with the standard climate dumping frequency of 10-days.
CRUN-CRUN tests (MONSooN)
For this test the model is run a second time as NRUN-CRUN jobsteps. The 2-day dump from the 1st CRUN test is then compared with this newly generated 2-day dump from the 2nd CRUN. This test bit-compares.
COMPARE - SUMMARY MODE ----------------------- Number of fields in file 1 = 14272 Number of fields in file 2 = 14272 Number of fields compared = 14272 FIXED LENGTH HEADER: Number of differences = 3 INTEGER HEADER: Number of differences = 0 REAL HEADER: Number of differences = 0 LEVEL DEPENDENT CONSTANTS: Number of differences = 0 LOOKUP: Number of differences = 0 DATA FIELDS: Number of fields with differences = 0 files compare, ignoring Fixed Length Header
This means that it is possible to re-run a UKCA job-step, and assuming that the dump frequency is the same (and that they started from the same dump), then the results will be reproducible.
change of dump frequency test (MONSooN)
In this test a 2-day long run is performed with daily dumping (in a single job-step), and then a 2-day run is performed with 2-day dumping. In this case, these dumps DO NOT bit-compare.
COMPARE - SUMMARY MODE ----------------------- Number of fields in file 1 = 14272 Number of fields in file 2 = 14272 Number of fields compared = 14272 FIXED LENGTH HEADER: Number of differences = 3 INTEGER HEADER: Number of differences = 0 REAL HEADER: Number of differences = 3 LEVEL DEPENDENT CONSTANTS: Number of differences = 0 LOOKUP: Number of differences = 0 DATA FIELDS: Number of fields with differences = 12221 Field 1 : Stash Code 2 : U COMPNT OF WIND AFTER TIMESTEP : Number of differences = 27840 ... ... Field 14076 : Stash Code 38405 : DRY PARTICLE DIAMETER AITKEN-INS : Number of differences = 68 files DO NOT compare
This may also be to do with the way certain fields are initialised in UKCA. You should therefore try to maintain the 10-day dumping frequency. If this is done, and the job-steps are consistently a month long, then a repeated run of this configuration will be bit-reproducible, as the CRUN-CRUN test was passed.
This should be compared to the equivalent test performed on ARCHER, which passed.
Initial conditions and forcing
xkawawas initialised from the final dump ofxjcin(dated 2015-12-01), then re-dated usingchange_dump_dateto 1999-12-01.- See
/projects/ukca/inputs/initial/vn84GA4_UKCA.19991201_00.txt
- See
- SSTs and sea-ice use daily values from the Reynolds dataset, produced by meaning over the transient values from 1995-01-01 to 2005-01-01 using
cdo ydaymean.- See
/projects/ukca/inputs/ancil/surf/reynolds.qrclim.sst.avg2000.txtfor SSTs. - See
/projects/ukca/inputs/ancil/surf/reynolds.qrclim.seaice.avg2000.txtfor the sea-ice. - Note: there was a minor error with these files, in that the time is set to midnight and not noon. However, this is unlikely to cause any major problems.
- See
- Time-slice conditions for the year 2000 (actual date 2000-12-01), known as TS2000, were used using the RCP scenario conditions for GHGs and WMO2011 values for ODSs.
- These are the forcings specified for the CCMI transient REF-C1/2 simulations
- For further information, see the CCMI website and the RCP database
- These values are set in the Spec of trace gases section of the UMUI
- Note that CFC-114 is only used in UKCA and not in the radiation scheme (as the spectral file does not consider CFC-114). This is done with the hand-edit
~ukca/hand_edits/VN8.4/CFC-114_not_in_Rad.ed.
- Note that CFC-114 is only used in UKCA and not in the radiation scheme (as the spectral file does not consider CFC-114). This is done with the hand-edit
- These were calculated using the
scenariofunction on PUMA:
- These are the forcings specified for the CCMI transient REF-C1/2 simulations
$ /home/ukca/bin/scenario 2000/12/01 USER USER-SUPPLIED SCENARIO SELECTED. PLEASE INPUT FILENAME /home/ukca/tools/scenario/RCP6_MIDYR_CONC_WMO2011_CCMI.DAT ----------------------------------------------- | 2000/12/01 USER SCENARIO: | ----------------------------------------------- | CFCl3 = 1.23542E-09 CFC11/F11 | | CF2Cl2 = 2.26638E-09 CFC12/F12 | | CF2ClCFCl2 = 5.29666E-10 CFC113/F113 | | CF2ClCF2Cl = 9.73663E-11 CFC114/F114 | | CF2ClCF3 = 4.25934E-11 CFC115/F115 | | CCl4 = 5.19557E-10 | | MeCCl3 = 1.95889E-10 CH3CCl3 | | CHF2Cl = 4.31321E-10 HCFC22 | | MeCFCl2 = 5.35084E-11 HCFC141b | | MeCF2Cl = 4.24961E-11 HCFC142b | | CF2ClBr = 2.34011E-11 H1211 | | CF2Br2 = 2.74854E-13 H1202 | | CF3Br = 1.46645E-11 H1301 | | CF2BrCF2Br = 4.48740E-12 H2402 | | MeCl = 9.58777E-10 CH3Cl | | MeBr = 2.80100E-11 CH3Br | | CH2Br2 = 1.80186E-11 | | N2O = 4.80116E-07 | | CH4 = 9.67017E-07 | | CF3CHF2 = 6.18271E-12 HFC125 | | CH2FCF3 = 5.43855E-11 HFC134a | | H2 = 3.45280E-08 | | N2 = 7.54682E-01 | | CO2 = 5.61246E-04 | ----------------------------------------------- UM/UKCA LBC MMRs for: 2000/12/01, using the USER scenario VALUES FOR USE IN THE UMUI (ZERO VALUES CAN BE TREATED AS "Excluded"): CH4 = 9.670E-07 N2O = 4.801E-07 CFC11 = 1.235E-09 CFC12 = 2.266E-09 CFC113 = 5.297E-10 HCFC22 = 4.313E-10 HFC125 = 6.183E-12 HFC134a = 5.439E-11 CO2 = 5.61246E-04 VALUES FOR USE IN THE UKCA HAND-EDIT: MeBrMMR=2.80100E-11, MeClMMR=9.58777E-10, CH2Br2MMR=1.80186E-11, H2MMR=3.45280E-08, N2MMR=0.75468 , CFC114MMR=9.73663E-11, CFC115MMR=4.25934E-11, CCl4MMR=5.19557E-10, MeCCl3MMR=1.95889E-10, HCFC141bMMR=5.35084E-11, HCFC142bMMR=4.24961E-11, H1211MMR=2.34011E-11, H1202MMR=2.74854E-13, H1301MMR=1.46645E-11, H2402MMR=4.48740E-12,
- The following sources are used for the chemistry and aerosol emissions:
- The 2D Sulphur-Cycle Emissions are the year 2000 values extracted from the standard CMIP5 dataset used for CLASSIC, which can be found at
/projects/um1/ancil/atmos/n96/classic_aerosol/cmip5/1970_2010/v0/qrclim.sulpsurf.
- Year 2000 AR5 emissions are used for NO (s0i301), CO (s0i303), HCHO (s0i304), C2H6 (s0i305), C3H8 (s0i306), (CH3)2CO (s0i307; Me2CO), CH3CHO (s0i308) MeCHO), Black Carbon (BC) fossil fuel surface emissions (s0i310), Black Carbon (BC) biofuel surface emissions (s0i311), Organic Carbon (OC) fossil fuel surface emissions (s0i312), Organic Carbon (OC) biofuel surface emissions (s0i313), and 3D NO aircraft emissions (s0i340).
- Year 2000 GEIA emissions are used for C5H8 (s0i309; isoprene) and Monoterpenes (s0i314).
- Year 2005 MEGAN emissions are used for CH3OH, labelled as NVOC in the STASHmaster file (s0i315; MeOH).
- http://tropo.aeronomie.be/models/methanol.htm
- Note: 2005 emissions were used for consistency with comparing to a vn7.3 CheST job - see the information on the NOy PEG.
- Year 2000 GFED2 emissions are used for 3D Black Carbon (s0i322; BC) and 3D Organic Carbon (s0i323; OC).
- The 2D Sulphur-Cycle Emissions are the year 2000 values extracted from the standard CMIP5 dataset used for CLASSIC, which can be found at
Making a transient run from a timeslice run
As noted above, this job is configured to run as a timeslice. It is relatively straight-forward to change this job to run as a timeslice, although you will need to change the initial conditions of the long-lived chemical tracers.
You will need to:
- Obtain or create a set of SST and sea-ice ancillaries for the time-period you are interested in.
- Obtain or create a set of emissions ancillaries for the time-period you are interested in.
It is possible to use a start-dump from a timeslice job to initialise a transient run. However, care must be taken over the values for the long-lived chemical tracers, which will have lower boundary conditions set for their surface concentrations. As described above, the UMUI is used to set the values for these gases. It is possible to use the UKCA routine ukca_rcp_scenario to read the RCP forcing files produced for CMIP5, and an example hand-edit is
$ cat /home/ukca/hand_edits/VN8.4/UKCA_RCP6.0.ed # UKCA_RCP6.0 # (vn8.5) # control variables to tell the model to use the RCP6.0 scenario, and # where the data for this is located. # # NOTE: FOR THIS HAND-EDIT TO WORK, THE OPTION 'OVERRIDE DEFAULTS' MUST # BE SET IN THE UMUI ed CNTLATM <<\EOF1 /L_UKCA_USEUMUIVALS/ d i I_UKCA_SCENARIO=2, UKCA_RCPdir='/projects/ukca/nlabra/scenario' UKCA_RCPfile='RCP6_MIDYR_CONC.DAT' . wq EOF1
However, this only affects the values in UKCA - the values seen by the radiation will not be affected. You must therefore either make equivalent changes to the UMUI panels which specify these trace gases, or you will need to include additional Fortran code in atmos_physics1.F90 (please contact Luke Abraham for more information on this option).
As well as changing how the lower boundary condition for these gases is specified, it is advisable to re-scale the gases according to their new surface concentrations. As the CheST chemistry scheme lumps the Cl contribution into CFC11 and CFC12, and the Br contribution into CH3Br (MeBr), it is not easy to use the scenario routine described above to calculate these concentrations. However, there is a lumped scenario function, which can be found at ~ukca/bin/scenario.lumped which will produce these values, and is used in the same way, e.g.
$ ~ukca/bin/scenario.lumped 2000/12/01 USER USER-SUPPLIED SCENARIO SELECTED. PLEASE INPUT FILENAME /home/ukca/tools/scenario/RCP6_MIDYR_CONC_WMO2011_CCMI.DAT ------------------------------------------------------- | 2000/12/01 USER SCENARIO (LUMPED MMR): | ------------------------------------------------------- | CFCl3 = 2.99423E-09 CFC11/F11 (LUMPED) | | CF2Cl2 = 3.16674E-09 CFC12/F12 (LUMPED) | | MeBr = 7.07330E-11 CH3Br (LUMPED) | | N2O = 4.80116E-07 | | CH4 = 9.67017E-07 | | H2 = 3.45280E-08 | | COS = 5.20000E-10 | -------------------------------------------------------
You should run this routine for the date you wish to start on, and compare the values to those above (or, if applying this method to a different timeslice run, to the values generated by the scenario.lumped program for the date of that timeslice) - any differences will mean that the fields will need to be rescaled (by default, H2 and COS are actually constant). This can be easily done using the climate data operators (by first using Xconv to extract these to netCDF from the dump you wish to use). You will also need to subtract the CFC11/CFC12 and CH3Br contributions from the LUMPED Cl (s34i100) and LUMPED Br (s34i099) tracers. To do this, do:
- Extract your CFC11 (s34i055), CFC12 (s34i056), CH3Br (s34i057), LUMPED Br (s34i099; assumed to be as BrO), and LUMPED Cl (s34i100; assumed to be as HCl) tracers from the dump file.
- Rescale your CFC11, CFC12, and CH3Br fields to the correct values as calculated from the
~ukca/bin/scenario.lumpedprogram (i.e. multiply by the factornew/original). - Extract the CH4 (s34i009) and N2O (s34i049) fields from the dump file and rescale these in the same way.
- Convert all Cl and Br fields to vmr. The required conversion factors can be found in the
src/atmosphere/UKCA/ukca_constants.F90. You should divide the tracer concentration by the c_species value for that species. Note that you should use the values for BrO for LUMPED Br and HCl for LUMPED Cl. - Subtract the original CFC11 and CFC12 fields (in vmr) from the LUMPED Cl vmr field, and then add-in the new CFC11 and CFC12 fields (also in vmr).
- Subtract the original CH3Br field (in vmr) from the LUMPED Br vmr field, and then add-in the new CH3Br field (also in vmr).
- Convert your new LUMPED Cl to mmr (using the conversion factor for HCl, as before).
- Convert your new LUMPED Br to mmr (using the conversion factor for BrO, as before).
- Use Xancil to convert these netCDF fields to ancillary file format (use the generalised ancillary file option).
- In the UMUI, turn on reconfiguration, and in the Initialisation of user prognostics panel use option 7 to point to the ancillary file containing these fields.
Note: once you have made these changes, you should spin the model up again. We suggest at least a 10-year spin-up. This can either be done as another timeslice, or as a transient run started earlier than needed, e.g. for a 1960-2010 simulation (starting at 1959-12-01) you could either
- Start the run in 1949-12-01 and run for 10 years to produce the 1959-12-01 start dump.
- Make a TS1960 timeslice (with forcing values set to e.g. 1959-12-01) and run this for 10-years to get a 1959-12-01 start dump. In this case you could also start the model from 1949-12-01, but would have the SSTs, sea-ice, and emissions etc. fixed at 1959 values.
ARCHER port: xkawe
This job has been ported to ARCHER as job xkawe, which included the updates to the FCM branch to revision number r16246, as stated above. More information about ARCHER can be found at www.archer.ac.uk.
Ported Model
The development of this job is very similar to xkawa as described in the base model section above, but branches off from job xjcim like so:
xjcim (made TS2000 - see Initial conditions and forcing) owned by Luke Abraham
xjnjb (ARCHER port of xjcim, with some bugfixes) owned by Karthee Sivalingam
xjqka (UKCA Tutorials base job; some branch consolidation and use of pre-compiled builds) owned by Luke Abraham
xjrna (direct copy of xjqka) owned by ukca UMUI user
xkawe (code changes etc. as per xkawf - i.e. bugfixes at r16485) owned by Luke Abraham
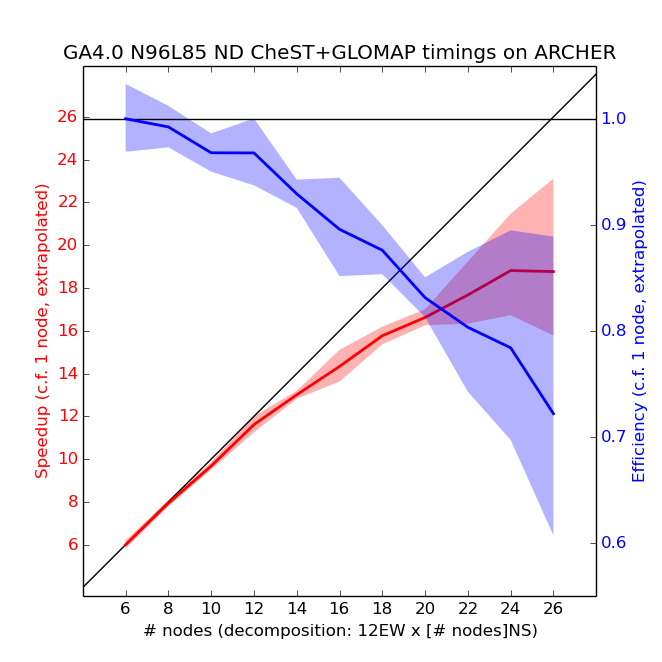
Scaling (ARCHER)
Each compute node contains two 2.7 GHz, 12-core Ivy Bridge processors, which can support 2 hardware threads, and there is 64GB of RAM per node. Due to memory restrictions, UKCA is unable to run on less than 6 nodes (meaning that the 4-node debug queue is not available). As noted below, the EW domain decomposition needed to be a multiple of 12. All simulations used 2 OpenMP threads with 12 MPI tasks per node (halving the number of cores available per node). Only 12EW decomposition was used, as the scaling tests on MONSooN showed that this was preferable over the 24EW decomposition. Also, Karthee Sivalingam noticed that 24 MPI tasks per node was not stable for jobs longer than 12 hours due to memory issues when he ported over the ARCHER job xjnjb (a copy of xjcim).
Scaling tests have been done from 6 to 26 nodes of ARCHER with a series of 1-day runs, with the results presented below.
The speedup in the plot above is calculated by assuming a linear scaling from 6 nodes down to 1 node. 5 simulations were performed for each number of nodes, and the envelope is 2 standard deviations (assuming that the standard deviation of the extrapolated 1-node data-point is the mean of the standard deviations of all other points). From these tests the recommended decomposition is 12 EW x 12 NS (i.e. 12 nodes, or 144 cores using 12 MPI tasks per node), although any number of nodes from 6-12 should cost approximately the same amount of allocation units as there is linear scaling. Running on 12 nodes means that the model will complete 1 model month in approximately 2 hours. Above 12 nodes the model will still run quicker, but as the scaling is no longer linear this will 'cost' more per model year. The model should not be run on more than 24 nodes, as the model will then become slower than if run in a 12EWx24NS decomposition. There are 3008 nodes (72,192 cores) on ARCHER, and so running on more nodes (i.e 12 rather than 6) should not significantly impact queue time.
Also note that the model run length has a much larger standard deviation ("jitter") on ARCHER than on MONSooN. Users should ensure that the requested run-time is at least 10% longer than the estimated run-time to ensure that the model does not exceed this limit due to this jitter.
Using the ARCHER kAU calculator this would give around 115kAU per model year (a notional cost of £90.85 per model year), including the 10% jitter.
cumf tests (ARCHER)
As was done for MONSooN, cumf tests were also done on ARCHER. The utility can be found at /work/n02/n02/hum/vn8.4/cce/utils/cumf.
Note: A change of domain decomposition test was not performed as it is known that this configuration of UKCA will fail this test. This is due to the way that the Newton Raphson chemical solver converges, which is done over the whole domain. While the results would be different across different domain decompositions, they are all scientifically valid. So long as the decomposition is not changed during a run, results will be comparable (also taking the results of the other tests into account).
All tests were run without STASH, climate meaning, or the UKCA evaluation suite hand-edit (~mdalvi/umui_jobs/hand_edits/vn8.4/add_ukca_eval1_diags_l85.ed), as all of these can affect the dump by placing temporary fields in it. The model was run for 2 days, either with daily or 2-day dumping, and for the NRUN-CRUN test, the first step was for 1 day followed by a new job step for the 2nd day. Reconfiguration was only run once, at the start of the 1st test, and after that point the same .astart file was used by all jobs.
The following cumf tests were performed using revision r16246. It is not anticipated that the changes in r16485 will change these.
NRUN-NRUN tests (ARCHER)
For this test the model is run twice. The 2-day dumps are then compared. For this test it makes no difference if you compare the dumps produced using daily-dumping or 2-day dumping, as both pass this test (when compared to the equivalent dump produced using the same dumping frequency).
COMPARE - SUMMARY MODE ----------------------- Number of fields in file 1 = 14272 Number of fields in file 2 = 14272 Number of fields compared = 14272 FIXED LENGTH HEADER: Number of differences = 2 INTEGER HEADER: Number of differences = 0 REAL HEADER: Number of differences = 0 LEVEL DEPENDENT CONSTANTS: Number of differences = 0 LOOKUP: Number of differences = 0 DATA FIELDS: Number of fields with differences = 0 files compare, ignoring Fixed Length Header
NRUN-CRUN tests (ARCHER)
For this test the 2nd day dump of a daily dumping run (which has been run in a single jobstep) is compared with the 2nd day dump of a run where this was produced on a CRUN step (i.e. where the 1st day dump was produced on the NRUN step). In this case, the dumps DO NOT compare.
COMPARE - SUMMARY MODE ----------------------- Number of fields in file 1 = 14272 Number of fields in file 2 = 14272 Number of fields compared = 14272 FIXED LENGTH HEADER: Number of differences = 3 INTEGER HEADER: Number of differences = 0 REAL HEADER: Number of differences = 3 LEVEL DEPENDENT CONSTANTS: Number of differences = 0 LOOKUP: Number of differences = 0 DATA FIELDS: Number of fields with differences = 12221 Field 1 : Stash Code 2 : U COMPNT OF WIND AFTER TIMESTEP : Number of differences = 27840 ... ... Field 14076 : Stash Code 38405 : DRY PARTICLE DIAMETER AITKEN-INS : Number of differences = 69 files DO NOT compare
This is not unexpected for UKCA, as there are many variables which are initialised at the start of a run, but not saved to a dump to be re-initialised correctly. This is a feature which will need to be addressed in the future.
It should be noted that this does not invalidate a run, or prevent you from re-running to fill-in data gaps. You should, however, ensure that you maintain the original job-step length. On MONSooN is it recommended that maintain a 1-month jobstep length, with the standard climate dumping frequency of 10-days.
CRUN-CRUN tests (ARCHER)
For this test the model is run a second time as NRUN-CRUN jobsteps. The 2-day dump from the 1st CRUN test is then compared with this newly generated 2-day dump from the 2nd CRUN. This test bit-compares.
COMPARE - SUMMARY MODE ----------------------- Number of fields in file 1 = 14272 Number of fields in file 2 = 14272 Number of fields compared = 14272 FIXED LENGTH HEADER: Number of differences = 3 INTEGER HEADER: Number of differences = 0 REAL HEADER: Number of differences = 0 LEVEL DEPENDENT CONSTANTS: Number of differences = 0 LOOKUP: Number of differences = 0 DATA FIELDS: Number of fields with differences = 0 files compare, ignoring Fixed Length Header
This means that it is possible to re-run a UKCA job-step, and assuming that the dump frequency is the same (and that they started from the same dump), then the results will be reproducible.
change of dump frequency test (ARCHER)
In this test a 2-day long run is performed with daily dumping (in a single job-step), and then a 2-day run is performed with 2-day dumping. In this case, these dumps bit-compare.
COMPARE - SUMMARY MODE ----------------------- Number of fields in file 1 = 14272 Number of fields in file 2 = 14272 Number of fields compared = 14272 FIXED LENGTH HEADER: Number of differences = 3 INTEGER HEADER: Number of differences = 0 REAL HEADER: Number of differences = 0 LEVEL DEPENDENT CONSTANTS: Number of differences = 0 LOOKUP: Number of differences = 0 DATA FIELDS: Number of fields with differences = 0 files compare, ignoring Fixed Length Header
Compare this to MONSooN, where this test failed. This means that on ARCHER, if you need to change the dump frequency for some reason at some point during a run, the run will still bit-compare to a run where this was not done. However, you should still try to maintain the 10-day dumping frequency.
Known Issues
Interactive Dry Deposition Scheme
Currently it is possible to request dry-deposition for a species in the ukca_chem_strattrop.F90 module (the first 1 in the last three columns of numbers) e.g.
! 30 DD:12,WD:12, chch_t( 30,'HCl ', 1,'TR ',' ', 1, 1, 0), &
but if values for the required species have not been set in ukca_aerod.F90 and ukca_surfddr.F90 then no dry deposition will in fact be calculated. Please see UKCA Chemistry and Aerosol Tutorial 7 (Adding dry deposition of chemical species) for details as to how to add new values.
Because of this, you should see warning messages such as this in the .leave file:
? Warning Message: Surface resistance values not set for HCl ? Warning Message: Surface resistance values not set for HOCl ? Warning Message: Surface resistance values not set for HBr ? Warning Message: Surface resistance values not set for HOBr ? Warning Message: Surface resistance values not set for DMSO ? Warning Message: Surface resistance values not set for Monoterp ? Warning Message: Surface resistance values not set for Sec_Org
Currently none of the above species (HCl, HOCl, HBr, HOBr, DMSO, Monoterp, or Sec_Org) are in fact dry deposited in this job configuration.
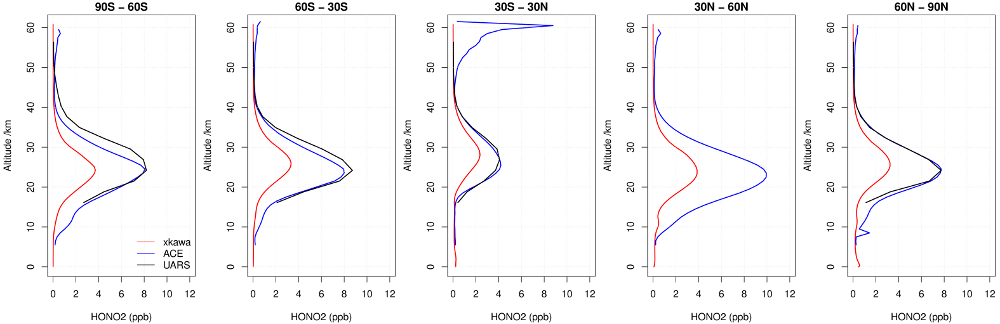
Low Stratospheric NOy
There is an issue with low stratospheric NOy in UKCA, as can be seen in the profiles of HNO3 above (calculated by meaning over the whole 10-year simulation.), and it is currently being investigated. For more information, see details from the NOy PEG.
High Stratospheric Sea-Salt Mixing Ratios
As Graham Mann notes below in the aerosol evaluation, there are high sea-salt mixing ratios in the stratosphere.
High Aerosol Optical Depth
As Jane Mulcahy notes below in the aerosol evaluation, this model configuration has high aerosol optical depth.
Reconfiguration Issues
Section 34, Item 163 (CLOUD DROPLET NO CONC^(-1/3) (m))
While field s34i163 does exist in the input dump, if reconfiguration is requested this will fail with an error unless an initial condition is supplied for this field. For more information see [1]. It is being investigated.
The file /projects/ukca/nlabra/ANCILS_N96L85/s34i163_Dec.anc contains the field from the initial dump (/projects/ukca/inputs/initial/vn84GA4_UKCA.19991201_00) and can be used in the Initialisation of user prognostics UMUI panel.
East-West Decomposition
On both MONSooN and ARCHER, the model is unable to run unless the East-West decomposition is a multiple of 12 (i.e. 12, 24 etc). This limits the possible domain decompositions, which must be multiples of 32 on MONSooN and 24 on ARCHER, to fit within the nodes efficiently.
MONSooN decomposition errors
On MONSooN, if running without multiple of 12 for the EW decomposition the job will exit with a floating point exception in glue_conv. The error message from ereport will be similar to:
???????????????????????????????????????????????????????????????????????????????? ???!!!???!!!???!!!???!!!???!!!???!!! ERROR ???!!!???!!!???!!!???!!!???!!!???!!!? ? Error in routine: glue_conv ? Error Code: 2 ? Error Message: Deep conv went to model top at point 8 in seg 1 on call 1 ? Error generated from processor: 100 ? This run generated 55 warnings ????????????????????????????????????????????????????????????????????????????????
ARCHER decomposition errors
On ARCHER, if running without a multiple of 12 for the EW decomposition the job will often exit with a segmentation fault in the ukca_calc_tropopause routine. This is the output when setting the ATP_ENABLED environment variable to 1 (set in Script Inserts and Modifications):
Application 9844563 is crashing. ATP analysis proceeding... ATP Stack walkback for Rank 19 starting: _start@start.S:113 __libc_start_main@libc-start.c:226 flumemain_@flumeMain.f90:48 um_shell_@um_shell.f90:1865 u_model_@u_model.f90:2931 ukca_main1_@ukca_main1-ukca_main1.f90:4848 ukca_calc_tropopause$ukca_tropopause_@ukca_tropopause.f90:178 ATP Stack walkback for Rank 19 done Process died with signal 11: 'Segmentation fault' Forcing core dumps of ranks 19, 0
Failing NRUN-CRUN tests
See the explanation of this test for MONSooN or ARCHER. The UKCA code management group is aware of this.
It should be noted that on MONSooN this configuration also fails the change of dump frequency test, whereas this is passed on ARCHER.
Diagnostics
ARCHER
As mentioned below, certain diagnostics (s30i310-316) could not be used with climate meaning, although the cause is uncertain. The traceback was
ATP Stack walkback for Rank 0 starting: _start@start.S:113 __libc_start_main@libc-start.c:226 flumemain_@flumeMain.f90:48 um_shell_@um_shell.f90:1865 u_model_@u_model.f90:3730 meanctl_@meanctl.f90:3631 acumps_@acumps.f90:1475 general_scatter_field_@general_scatter_field.f90:1098 stash_scatter_field_@stash_scatter_field.f90:955 gcg_ralltoalle_@gcg_ralltoalle.f90:180 gcg__ralltoalle_multi_@gcg_ralltoalle_multi.f90:335 ATP Stack walkback for Rank 0 done Process died with signal 11: 'Segmentation fault' Forcing core dumps of ranks 0, 1, 12, 13, 97, 140
This is solved by sending these diagnostics (and also s30i201-207 and s30i301) to the UPB stream.
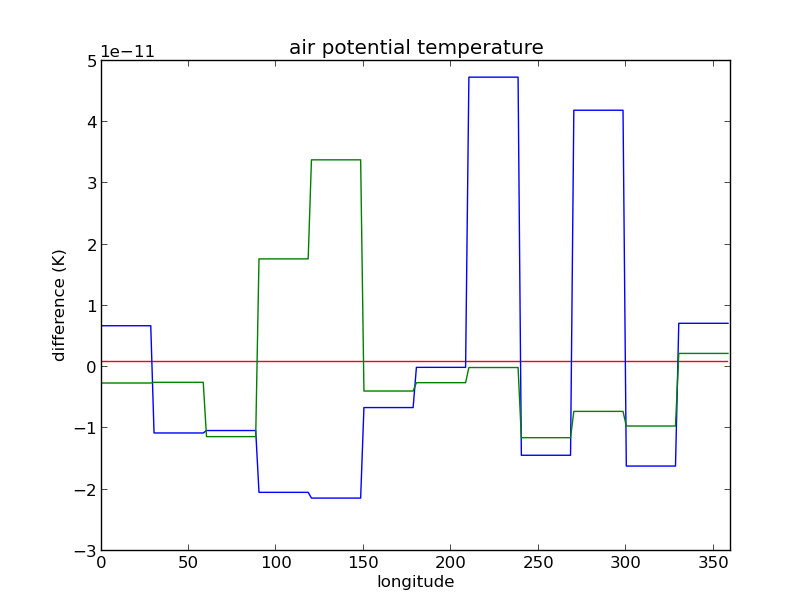
Non-uniform polar values for air potential temperature (theta)
Peter Uhe of CSIRO found that the values on the polar rows of the air potential temperature field (theta, STASH code m01s00i004) are non-uniform. This causes the model to crash on their systems.
The image above shows this problem. The blue and green lines have a different value of theta on each domain, whereas the red line does not (the lines are deviations from the mean).
The problem is also probably responsible for the inability of the model to run on EW decomposition other than multiples of 12.
Further Work Needed
Further work will need to be done to:
- Link the GLOMAP-mode aerosol to the heterogeneous reactions, especially for the stratospheric chemistry.
- Link the GLOMAP-mode aerosol to the Fast-JX interactive photolysis scheme.
- Extend the RCP scenario code to
atmos_physics1.F90to all the values of CO2, CH4, N2O etc. seen by the radiation scheme to be updated in the same way as for the chemistry. - Tidy-up the AerChem chemistry extensions required for GLOMAP-mode, so that they match better those for the TropIsop/CheT scheme.
- Link to the JULES land-surface scheme to allow for interactive isoprene and monoterpene emissions.
Results (MONSooN)
MOOSE
All results from this run were saved to MOOSE, and can be found at moose:/crum/xkawa. As well as the standard pp-output, monthly, seasonal, and annual supermeans were created and are also available in the ama.pp directory.
Running the command moo ls -l moose:/crum/xkawa gives
C colin.johnson 45.65 GBP 421803130880 2014-07-20 18:27:13 GMT moose:/crum/xkawa/ada.file C colin.johnson 3.09 GBP 28512510160 2014-07-24 15:56:40 GMT moose:/crum/xkawa/ama.pp C colin.johnson 20.50 GBP 189410089848 2014-07-25 11:20:50 GMT moose:/crum/xkawa/apa.pp C colin.johnson 24.85 GBP 229627024000 2014-07-25 11:22:33 GMT moose:/crum/xkawa/apb.pp C colin.johnson 20.95 GBP 193541722824 2014-07-20 20:25:13 GMT moose:/crum/xkawa/apm.pp C colin.johnson 6.93 GBP 63989055776 2014-07-20 18:31:17 GMT moose:/crum/xkawa/aps.pp C colin.johnson 1.73 GBP 16005498776 2014-07-20 18:32:26 GMT moose:/crum/xkawa/apy.pp
Further information on how to use MOOSE can be found on the collaboration twiki.
Note: If you take a copy of this job and run it, you must first manually make the MOOSE set to hold the data in the archive. This is done by
moo mkset --project-owner=project-YOUR_MONSooN_PROJECT -v moose:/crum/jobid
For instance, if you were in the UKCA project you would have --project-owner=project-ukca. This can be done after the job has started running. The archiving intelligently knows which files need archiving through the use of files named archive_XXXXXX.do. The files also control the deleting of files and dumps once they have been archived or are no longer needed. If, for some reason, the files cannot be archived (e.g. MOOSE is down or the set has not yet been made) then the files will not be deleted. They will continue being generated and existing on the /projects disk until they can be archived.
You should not need to do anything with the fieldsfiles until the whole simulation has been completed (in this case, the whole 10-years). When it does you will find that, while the climate mean files and dumps have been archived, the last files in the e.g. *.pa* or *.pb* streams etc will not have been. You will need to archive these manually by, e.g.
moo put -f -vv -c=umpp jobida.pzYYYYmmm moose:/crum/jobid/apz.pp/jobida.pzYYYYmmm.pp
Where the -c=umpp converts the files from 64-bit fieldsfiles to 32-bit pp-files. Remember to put the .pp at the end of the name of the file in the set on MOOSE.
Evaluation Suite Output (MONSooN)
A set of standard results from a mean of the 10-year run, as well as from each of the 10 years can be found in the following documents.
These were generated from the UKCA Evaluation Suite available on the MONSooN post-processor.
Mean over the 10-year simulation
Results from individual years
 Xkawa_EvalChemAer_yr01.pdf
Xkawa_EvalChemAer_yr01.pdf 
 Xkawa_EvalChemAer_yr02.pdf
Xkawa_EvalChemAer_yr02.pdf 
 Xkawa_EvalChemAer_yr03.pdf
Xkawa_EvalChemAer_yr03.pdf 
 Xkawa_EvalChemAer_yr04.pdf
Xkawa_EvalChemAer_yr04.pdf 
 Xkawa_EvalChemAer_yr05.pdf
Xkawa_EvalChemAer_yr05.pdf 
 Xkawa_EvalChemAer_yr06.pdf
Xkawa_EvalChemAer_yr06.pdf 
 Xkawa_EvalChemAer_yr07.pdf
Xkawa_EvalChemAer_yr07.pdf 
 Xkawa_EvalChemAer_yr08.pdf
Xkawa_EvalChemAer_yr08.pdf 
 Xkawa_EvalChemAer_yr09.pdf
Xkawa_EvalChemAer_yr09.pdf 
 Xkawa_EvalChemAer_yr10.pdf
Xkawa_EvalChemAer_yr10.pdf 
Aerosol Evaluation
Graham Mann has kindly produced the following plots for GLOMAP-mode evaluation:
Graham commented that: "The only issue I'd say is that there seems to be a problem with an anomalously high sea-salt mixing ratio in the stratosphere in these runs."
Jane Mulcahy has calculated the aerosol optical depth from the GLOMAP-mode fields, and has provided the following plots:
Jane has commented: "In my opinion the AODs are looking quite high particularly in spring and summer over anthropogenic regions. This is symptomatic of problems we are also seeing in our latest GA6 based runs. Looking at the surface sulphate and SO2 evaluation in Grahams plots this also looks high over Europe. So it is possible that there is insufficient removal of SO2 (no gas phase plume scavenging as yet). (The) biomass burning is also looking low in JJA. (I) would also caution users about high AOD."
Lightning NOx
The average annual Lightning NOx emitted is 4.03285 Tg(N)/year over the whole 10-year simulation.
The lightning NOx emitted in each of the individual years of the run is:
- year 01 4.06726 Tg(N)/year
- year 02 4.01840 Tg(N)/year
- year 03 4.06754 Tg(N)/year
- year 04 4.08320 Tg(N)/year
- year 05 3.99700 Tg(N)/year
- year 06 3.96347 Tg(N)/year
- year 07 4.01116 Tg(N)/year
- year 08 4.03512 Tg(N)/year
- year 09 4.04277 Tg(N)/year
- year 10 4.04452 Tg(N)/year
STASH Table
Below is a listing of all the STASH requests that were output by the model.
The output was sent to three different usage profiles, corresponding to:
- UPA: These files contain daily output, and go to the
*.pa*.ppfiles, held in theapa.ppdirectory on MOOSE. - UPB: These files contain monthly out from UKCA that could not fit in the climate meaning stream due to space limitations. These go to the
*.pb*.ppfiles, held in theapb.ppdirectory on MOOSE. - UPMEAN: This is the climate meaning stream, and holds a large number of dynamical and UKCA related diagnostics. These go to the:
*.pm*.ppfiles for monthly means, held in theapm.ppdirectory on MOOSE.*.ps*.ppfiles for seasonal means, held in theaps.ppdirectory on MOOSE.*.py*.ppfiles for annual means, held in theapy.ppdirectory on MOOSE.- no decadal means were created during this run, but if they were, they would be
*.px*.ppfiles, held in theapx.ppdirectory on MOOSE.
Running with STASH (including the use of the UKCA evaluation suite hand-edit ~mdalvi/umui_jobs/hand_edits/vn8.4/add_ukca_eval1_diags_l85.ed) increases the model run-time by 10-12%.
| Section | Item | Name | Time Profile | Domain Profile | Usage Profile |
| 0 | 4 | THETA AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 0 | 10 | SPECIFIC HUMIDITY AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 0 | 12 | QCF AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 0 | 23 | SNOW AMOUNT OVER LAND AFT TSTP KG/M | TDMPMN | DIAG | UPMEAN |
| 0 | 24 | SURFACE TEMPERATURE AFTER TIMESTEP | TDAYM | DIAG | UPA |
| 0 | 24 | SURFACE TEMPERATURE AFTER TIMESTEP | TDMPMN | DIAG | UPMEAN |
| 0 | 25 | BOUNDARY LAYER DEPTH AFTER TIMESTEP | TDMPMN | DIAG | UPMEAN |
| 0 | 28 | SURFACE ZONAL CURRENT AFTER TIMESTE | TDMPMN | DIAG | UPMEAN |
| 0 | 29 | SURFACE MERID CURRENT AFTER TIMESTE | TDMPMN | DIAG | UPMEAN |
| 0 | 31 | FRAC OF SEA ICE IN SEA AFTER TSTEP | TDMPMN | DIAG | UPMEAN |
| 0 | 32 | SEA ICE DEPTH (MEAN OVER ICE) | TDMPMN | DIAG | UPMEAN |
| 0 | 58 | SULPHUR DIOXIDE EMISSIONS | TDMPMN | DIAG | UPMEAN |
| 0 | 59 | DIMETHYL SULPHIDE EMISSIONS (ANCIL) | TDMPMN | DIAG | UPMEAN |
| 0 | 101 | SO2 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 0 | 102 | DIMETHYL SULPHIDE MIX RAT AFTER TS | TDMPMN | DALLTH | UPMEAN |
| 0 | 103 | SO4 AITKEN MODE AEROSOL AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 0 | 104 | SO4 ACCUM. MODE AEROSOL AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 0 | 105 | SO4 DISSOLVED AEROSOL AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 0 | 121 | 3D NATURAL SO2 EMISSIONS KG/M2/S | TDMPMN | DALLTH | UPMEAN |
| 0 | 126 | HIGH LEVEL SO2 EMISSIONS KG/M2/S | TDMPMN | DIAG | UPMEAN |
| 0 | 132 | DMS CONCENTRATION IN SEAWATER | TDMPMN | DIAG | UPMEAN |
| 0 | 211 | CCA WITH ANVIL AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 0 | 254 | QCL AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 0 | 266 | BULK CLOUD FRACTION IN EACH LAYER | TDMPMN | DALLTH | UPMEAN |
| 0 | 301 | NOx surf emissions | TDMPUKC3 | DIAG | UPMEAN |
| 0 | 302 | CH4 surf emissions | TDMPUKC3 | DIAG | UPMEAN |
| 0 | 303 | CO surf emissions | TDMPUKC3 | DIAG | UPMEAN |
| 0 | 309 | C5H8 surf emissions | TDMPUKC3 | DIAG | UPMEAN |
| 0 | 310 | BC fossil fuel surf emissions | TDMPUKC3 | DIAG | UPMEAN |
| 0 | 311 | BC biofuel surf emissions | TDMPUKC3 | DIAG | UPMEAN |
| 0 | 312 | OC fossil fuel surf emissions | TDMPUKC3 | DIAG | UPMEAN |
| 0 | 313 | OC biofuel surf emissions | TDMPUKC3 | DIAG | UPMEAN |
| 0 | 314 | Monoterpene surf emissions | TDMPUKC3 | DIAG | UPMEAN |
| 0 | 322 | BC BIOMASS 3D EMISSION | TDMPUKC3 | DALLTH | UPMEAN |
| 0 | 323 | OC BIOMASS 3D EMISSION | TDMPUKC3 | DALLTH | UPMEAN |
| 0 | 340 | NOX AIRCRAFT EMS IN KG/S/GRIDCELL | TDMPUKC3 | DALLTH | UPMEAN |
| 0 | 407 | PRESSURE AT RHO LEVELS AFTER TS | TDMPMN | DALLRH | UPMEAN |
| 0 | 408 | PRESSURE AT THETA LEVELS AFTER TS | TDMPMN | DALLTH | UPMEAN |
| 0 | 409 | SURFACE PRESSURE AFTER TIMESTEP | TDMPMN | DIAG | UPMEAN |
| 0 | 507 | OPEN SEA SURFACE TEMP AFTER TIMESTE | TDMPMN | DIAG | UPMEAN |
| 0 | 509 | SEA ICE ALBEDO AFTER TS | TDMPMN | DIAG | UPMEAN |
| 1 | 201 | NET DOWN SURFACE SW FLUX: SW TS ONL | TDMPMN | DIAG | UPMEAN |
| 1 | 203 | NET DN SW RAD FLUX:OPEN SEA:SEA MEA | TDMPMN | DIAG | UPMEAN |
| 1 | 207 | INCOMING SW RAD FLUX (TOA): ALL TSS | TDMPMN | DIAG | UPMEAN |
| 1 | 208 | OUTGOING SW RAD FLUX (TOA) | TDMPMN | DIAG | UPMEAN |
| 1 | 209 | CLEAR-SKY (II) UPWARD SW FLUX (TOA) | TDMPMN | DIAG | UPMEAN |
| 1 | 210 | CLEAR-SKY (II) DOWN SURFACE SW FLUX | TDMPMN | DIAG | UPMEAN |
| 1 | 211 | CLEAR-SKY (II) UP SURFACE SW FLUX | TDMPMN | DIAG | UPMEAN |
| 1 | 221 | LAYER CLD LIQ RE * LAYER CLD WEIGHT | TDMPMN | DALLTHCL | UPMEAN |
| 1 | 223 | LAYER CLOUD WEIGHT FOR MICROPHYSICS | TDMPMN | DALLTHCL | UPMEAN |
| 1 | 225 | CONV CLOUD LIQ RE * CONV CLD WEIGHT | TDMPMN | DALLTHCL | UPMEAN |
| 1 | 226 | CONV CLOUD WEIGHT FOR MICROPHYSICS | TDMPMN | DALLTHCL | UPMEAN |
| 1 | 235 | TOTAL DOWNWARD SURFACE SW FLUX | TDMPMN | DIAG | UPMEAN |
| 1 | 241 | DROPLET NUMBER CONC * LYR CLOUD WGT | TDMPMN | DALLTHCL | UPMEAN |
| 1 | 242 | LAYER CLOUD LWC * LAYER CLOUD WEIGH | TDMPMN | DALLTHCL | UPMEAN |
| 1 | 280 | COLUMN-INTEGRATED Nd * SAMP. WEIGHT | TDMPMN | DIAG | UPMEAN |
| 1 | 281 | SAMP. WEIGHT FOR COL. INT. Nd | TDMPMN | DIAG | UPMEAN |
| 1 | 282 | 2-D CLOUD-TOP CDNC x WEIGHT (cm-3) | TDMPMN | DIAG | UPMEAN |
| 1 | 283 | WEIGHT FOR 2-D CLOUD-TOP CDNC | TDMPMN | DIAG | UPMEAN |
| 2 | 201 | NET DOWN SURFACE LW RAD FLUX | TDMPMN | DIAG | UPMEAN |
| 2 | 203 | NET DN LW RAD FLUX:OPEN SEA:SEA MEA | TDMPMN | DIAG | UPMEAN |
| 2 | 204 | TOTAL CLOUD AMOUNT IN LW RADIATION | TDMPMN | DIAG | UPMEAN |
| 2 | 205 | OUTGOING LW RAD FLUX (TOA) | TDMPMN | DIAG | UPMEAN |
| 2 | 206 | CLEAR-SKY (II) UPWARD LW FLUX (TOA) | TDMPMN | DIAG | UPMEAN |
| 2 | 207 | DOWNWARD LW RAD FLUX: SURFACE | TDMPMN | DIAG | UPMEAN |
| 2 | 208 | CLEAR-SKY (II) DOWN SURFACE LW FLUX | TDMPMN | DIAG | UPMEAN |
| 2 | 251 | AITKEN MODE (SOLUBLE) STRATO AOD | TDAYM | DIAGAOT | UPA |
| 2 | 251 | AITKEN MODE (SOLUBLE) STRATO AOD | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 252 | ACCUM MODE (SOLUBLE) STRATO AOD | TDAYM | DIAGAOT | UPA |
| 2 | 252 | ACCUM MODE (SOLUBLE) STRATO AOD | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 253 | COARSE MODE (SOLUBLE) STRATO AOD | TDAYM | DIAGAOT | UPA |
| 2 | 253 | COARSE MODE (SOLUBLE) STRATO AOD | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 254 | AITKEN MODE (INSOL) STRATO AOD | TDAYM | DIAGAOT | UPA |
| 2 | 254 | AITKEN MODE (INSOL) STRATO AOD | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 255 | ACCUM MODE (INSOL) STRATO AOD | TDAYM | DIAGAOT | UPA |
| 2 | 255 | ACCUM MODE (INSOL) STRATO AOD | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 256 | COARSE MODE (INSOL) STRATO AOD | TDAYM | DIAGAOT | UPA |
| 2 | 256 | COARSE MODE (INSOL) STRATO AOD | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 284 | SULPHATE OPTICAL DEPTH IN RADIATION | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 285 | MINERAL DUST OPTICAL DEPTH IN RADN. | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 298 | TOTAL OPTICAL DEPTH IN RADIATION | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 300 | AITKEN MODE (SOLUBLE) OPTICAL DEPTH | TDAYM | DIAGAOT | UPA |
| 2 | 300 | AITKEN MODE (SOLUBLE) OPTICAL DEPTH | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 301 | ACCUM MODE (SOLUBLE) OPTICAL DEPTH | TDAYM | DIAGAOT | UPA |
| 2 | 301 | ACCUM MODE (SOLUBLE) OPTICAL DEPTH | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 302 | COARSE MODE (SOLUBLE) OPTICAL DEPTH | TDAYM | DIAGAOT | UPA |
| 2 | 302 | COARSE MODE (SOLUBLE) OPTICAL DEPTH | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 303 | AITKEN MODE (INSOL) OPTICAL DEPTH | TDAYM | DIAGAOT | UPA |
| 2 | 303 | AITKEN MODE (INSOL) OPTICAL DEPTH | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 304 | ACCUM MODE (INSOL) OPTICAL DEPTH | TDAYM | DIAGAOT | UPA |
| 2 | 304 | ACCUM MODE (INSOL) OPTICAL DEPTH | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 305 | COARSE MODE (INSOL) OPTICAL DEPTH | TDAYM | DIAGAOT | UPA |
| 2 | 305 | COARSE MODE (INSOL) OPTICAL DEPTH | TDMPMN | DIAGAOT | UPMEAN |
| 2 | 422 | DUST OPTICAL DEPTH FROM PROGNOSTIC | TDMPMN | DIAGAOT | UPMEAN |
| 3 | 217 | SURFACE SENSIBLE HEAT FLUX W/M | TDMPMN | DIAG | UPMEAN |
| 3 | 223 | SURFACE TOTAL MOISTURE FLUX KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 3 | 224 | WIND MIX EN'GY FL TO SEA:SEA MN W/M | TDMPMN | DIAG | UPMEAN |
| 3 | 225 | 10 METRE WIND U-COMP B GRID | TDMPMN | DIAG | UPMEAN |
| 3 | 226 | 10 METRE WIND V-COMP B GRID | TDMPMN | DIAG | UPMEAN |
| 3 | 232 | EVAP FROM OPEN SEA: SEA MEAN KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 3 | 234 | SURFACE LATENT HEAT FLUX W/M | TDMPMN | DIAG | UPMEAN |
| 3 | 237 | SPECIFIC HUMIDITY AT 1.5M | TDMPMN | DIAG | UPMEAN |
| 3 | 245 | RELATIVE HUMIDITY AT 1.5M | TDMPMN | DIAG | UPMEAN |
| 3 | 261 | GROSS PRIMARY PRODUCTIVITY KG C/M2/ | TDMPMN | DIAG | UPMEAN |
| 3 | 262 | NET PRIMARY PRODUCTIVITY KG C/M2/S | TDMPMN | DIAG | UPMEAN |
| 3 | 263 | PLANT RESPIRATION KG/M2/S | TDMPMN | DIAG | UPMEAN |
| 3 | 291 | NET PRIMARY PRODUCTIVITY ON PFTS | TDMPMN | DPFTS | UPMEAN |
| 3 | 293 | SOIL RESPIRATION KG C/M2/ | TDMPMN | DIAG | UPMEAN |
| 3 | 304 | TURBULENT MIXING HT AFTER B.LAYER m | TDMPMN | DIAG | UPMEAN |
| 3 | 305 | STABLE BL INDICATOR | TDMPMN | DIAG | UPMEAN |
| 3 | 307 | WELL_MIXED BL INDICATOR | TDMPMN | DIAG | UPMEAN |
| 3 | 310 | CUMULUS-CAPPED BL INDICATOR | TDMPMN | DIAG | UPMEAN |
| 3 | 332 | TOA OUTGOING LW RAD AFTER B.LAYER | TDMPMN | DIAG | UPMEAN |
| 3 | 392 | X-COMP OF MEAN SEA SURF STRESS N/M2 | TDMPMN | DIAG | UPMEAN |
| 3 | 394 | Y-COMP OF MEAN SEA SURF STRESS N/M2 | TDMPMN | DIAG | UPMEAN |
| 4 | 203 | LARGE SCALE RAINFALL RATE KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 4 | 204 | LARGE SCALE SNOWFALL RATE KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 4 | 216 | SO2 SCAVENGED BY LS PPN KG/M2/S | TDMPMN | DIAG | UPMEAN |
| 4 | 219 | SO4 DIS SCAVNGD BY LS PPN KG/M2/S | TDMPMN | DIAG | UPMEAN |
| 5 | 205 | CONVECTIVE RAINFALL RATE KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 5 | 206 | CONVECTIVE SNOWFALL RATE KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 5 | 214 | TOTAL RAINFALL RATE: LS+CONV KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 5 | 215 | TOTAL SNOWFALL RATE: LS+CONV KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 5 | 216 | TOTAL PRECIPITATION RATE KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 5 | 231 | CAPE TIMESCALE (DEEP) | TDMPMN | DIAG | UPMEAN |
| 5 | 238 | SO2 SCAVENGED BY CONV PPN KG/M2/SEC | TDMPMN | DIAG | UPMEAN |
| 5 | 239 | SO4 AIT SCAVNGD BY CONV PPN KG/M2/S | TDMPMN | DIAG | UPMEAN |
| 5 | 240 | SO4 ACC SCAVNGD BY CONV PPN KG/M2/S | TDMPMN | DIAG | UPMEAN |
| 5 | 241 | SO4 DIS SCAVNGD BY CONV PPN KG/M2/S | TDMPMN | DIAG | UPMEAN |
| 5 | 269 | DEEP CONVECTION INDICATOR | TDMPMN | DIAG | UPMEAN |
| 5 | 270 | SHALLOW CONVECTION INDICATOR | TDMPMN | DIAG | UPMEAN |
| 5 | 272 | MID LEVEL CONVECTION INDICATOR | TDMPMN | DIAG | UPMEAN |
| 5 | 277 | DEEP CONV PRECIP RATE KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 5 | 278 | SHALLOW CONV PRECIP RATE KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 5 | 279 | MID LEVEL CONV PRECIP RATE KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 5 | 320 | MASS FLUX DEEP CONVECTION | TDMPMN | D52TH | UPMEAN |
| 5 | 322 | MASS FLUX SHALLOW CONVECTION | TDMPMN | D52TH | UPMEAN |
| 5 | 323 | MASS FLUX MID-LEVEL CONVECTION | TDMPMN | D52TH | UPMEAN |
| 6 | 201 | X COMPONENT OF GRAVITY WAVE STRESS | TDMPMN | DALLTH | UPMEAN |
| 8 | 23 | SNOW MASS AFTER HYDROLOGY KG/M | TDMPMN | DIAG | UPMEAN |
| 8 | 208 | SOIL MOISTURE CONTENT | TDMPMN | DIAG | UPMEAN |
| 8 | 223 | SOIL MOISTURE CONTENT IN A LAYER | TDMPMN | DSOIL | UPMEAN |
| 8 | 225 | DEEP SOIL TEMP. AFTER HYDROLOGY DEG | TDMPMN | DSOIL | UPMEAN |
| 8 | 234 | SURFACE RUNOFF RATE KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 8 | 235 | SUB-SURFACE RUNOFF RATE KG/M2/ | TDMPMN | DIAG | UPMEAN |
| 16 | 4 | TEMPERATURE ON THETA LEVELS | TDMPMN | DALLTH | UPMEAN |
| 16 | 222 | PRESSURE AT MEAN SEA LEVEL | TDMPMN | DIAG | UPMEAN |
| 30 | 112 | WBIG Set to 1 if w GT 1.0m/s | TDMPMN | DALLTH | UPMEAN |
| 30 | 114 | WBIG Set to 1 if w GT 0.1m/s | TDMPMN | DALLTH | UPMEAN |
| 30 | 201 | U COMPNT OF WIND ON P LEV/UV GRID | TDAYM | DP36CCM | UPA |
| 30 | 201 | U COMPNT OF WIND ON P LEV/UV GRID | TDMPMN/TMONMN(*) | DP36CCM | UPMEAN/UPB(*) |
| 30 | 202 | V COMPNT OF WIND ON P LEV/UV GRID | TDAYM | DP36CCM | UPA |
| 30 | 202 | V COMPNT OF WIND ON P LEV/UV GRID | TDMPMN/TMONMN(*) | DP36CCM | UPMEAN/UPB(*) |
| 30 | 203 | W COMPNT OF WIND ON P LEV/UV GRID | TDAYM | DP36CCM | UPA |
| 30 | 203 | W COMPNT OF WIND ON P LEV/UV GRID | TDMPMN | DP17 | UPMEAN |
| 30 | 203 | W COMPNT OF WIND ON P LEV/UV GRID | TDMPMN/TMONMN(*) | DP36CCM | UPMEAN/UPB(*) |
| 30 | 204 | TEMPERATURE ON P LEV/UV GRID | TDAYM | DP36CCM | UPA |
| 30 | 204 | TEMPERATURE ON P LEV/UV GRID | TDMPMN/TMONMN(*) | DP36CCM | UPMEAN/UPB(*) |
| 30 | 205 | SPECIFIC HUMIDITY ON P LEV/UV GRID | TDAYM | DP36CCM | UPA |
| 30 | 205 | SPECIFIC HUMIDITY ON P LEV/UV GRID | TDMPMN/TMONMN(*) | DP36CCM | UPMEAN/UPB(*) |
| 30 | 206 | RELATIVE HUMIDITY ON P LEV/UV GRID | TDAYM | DP36CCM | UPA |
| 30 | 206 | RELATIVE HUMIDITY ON P LEV/UV GRID | TDMPMN/TMONMN(*) | DP36CCM | UPMEAN/UPB(*) |
| 30 | 207 | GEOPOTENTIAL HEIGHT ON P LEV/UV GRI | TDAYM | DP36CCM | UPA |
| 30 | 207 | GEOPOTENTIAL HEIGHT ON P LEV/UV GRI | TDMPMN/TMONMN(*) | DP36CCM | UPMEAN/UPB(*) |
| 30 | 208 | OMEGA ON P LEV/UV GRID | TDAYM | DP36CCM | UPA |
| 30 | 208 | OMEGA ON P LEV/UV GRID | TDMPMN | DP17 | UPMEAN |
| 30 | 208 | OMEGA ON P LEV/UV GRID | TDMPMN/TMONMN(*) | DP36CCM | UPMEAN/UPB(*) |
| 30 | 301 | HEAVYSIDE FN ON P LEV/UV GRID | TDAYM | DP36CCM | UPA |
| 30 | 301 | HEAVYSIDE FN ON P LEV/UV GRID | TDMPMN/TMONMN(*) | DP36CCM | UPMEAN/UPB(*) |
| 30 | 310 | RESIDUAL MN MERID. CIRC. VSTARBAR | TDAYM | DP36CCM | UPA |
| 30 | 310 | RESIDUAL MN MERID. CIRC. VSTARBAR | TDMPMN/not used(*) | DP36CCM | UPMEAN/not used(*) |
| 30 | 311 | RESIDUAL MN MERID. CIRC. WSTARBAR | TDAYM | DP36CCM | UPA |
| 30 | 311 | RESIDUAL MN MERID. CIRC. WSTARBAR | TDMPMN/not used(*) | DP36CCM | UPMEAN/not used(*) |
| 30 | 312 | ELIASSEN-PALM FLUX (MERID. COMPNT) | TDAYM | DP36CCM | UPA |
| 30 | 312 | ELIASSEN-PALM FLUX (MERID. COMPNT) | TDMPMN/not used(*) | DP36CCM | UPMEAN/not used(*) |
| 30 | 313 | ELIASSEN-PALM FLUX (VERT. COMPNT) | TDAYM | DP36CCM | UPA |
| 30 | 313 | ELIASSEN-PALM FLUX (VERT. COMPNT) | TDMPMN/not used(*) | DP36CCM | UPMEAN/not used(*) |
| 30 | 314 | DIVERGENCE OF ELIASSEN-PALM FLUX | TDAYM | DP36CCM | UPA |
| 30 | 314 | DIVERGENCE OF ELIASSEN-PALM FLUX | TDMPMN/not used(*) | DP36CCM | UPMEAN/not used(*) |
| 30 | 315 | MERIDIONAL HEAT FLUX | TDAYM | DP36CCM | UPA |
| 30 | 315 | MERIDIONAL HEAT FLUX | TDMPMN/not used(*) | DP36CCM | UPMEAN/not used(*) |
| 30 | 316 | MERIDIONAL MOMENTUM FLUX | TDAYM | DP36CCM | UPA |
| 30 | 316 | MERIDIONAL MOMENTUM FLUX | TDMPMN/not used(*) | DP36CCM | UPMEAN/not used(*) |
| 30 | 403 | TOTAL COLUMN DRY MASS RHO GRID | TDMPMN | DIAG | UPMEAN |
| 30 | 404 | TOTAL COLUMN WET MASS RHO GRID | TDMPMN | DIAG | UPMEAN |
| 30 | 405 | TOTAL COLUMN QCL RHO GRID | TDMPMN | DIAG | UPMEAN |
| 30 | 406 | TOTAL COLUMN QCF RHO GRID | TDMPMN | DIAG | UPMEAN |
| 30 | 419 | ENERGY CORR P GRID IN COLUMN W/M2 | TDMPMN | DIAG | UPMEAN |
| 30 | 451 | Pressure at Tropopause Level | TDMPMN | DIAG | UPMEAN |
| 30 | 452 | Temperature at Tropopause Level | TDMPMN | DIAG | UPMEAN |
| 30 | 453 | Height at Tropopause Level | TDMPMN | DIAG | UPMEAN |
| 34 | 1 | O3 MASS MIXING RATIO AFTER TIMESTEP | TDAYM | DALLTH | UPA |
| 34 | 1 | O3 MASS MIXING RATIO AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 2 | NO MASS MIXING RATIO AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 3 | NO3 MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 5 | N2O5 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 6 | HO2NO2 MASS MIXING RATIO AFTER TSTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 7 | HONO2 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 8 | H2O2 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 9 | CH4 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 10 | CO MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 11 | HCHO MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 12 | MeOOH MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 13 | HONO MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 14 | C2H6 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 15 | EtOOH MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 16 | MeCHO MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 17 | PAN MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 18 | C3H8 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 19 | n-PrOOH MASS MIXING RATIO AFTER TS | TDMPMN | DALLTH | UPMEAN |
| 34 | 20 | i-PrOOH MASS MIXING RATIO AFTER TS | TDMPMN | DALLTH | UPMEAN |
| 34 | 21 | EtCHO MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 22 | Me2CO MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 23 | MeCOCH2OOH MASS MIXING RATIO AFT TS | TDMPMN | DALLTH | UPMEAN |
| 34 | 24 | PPAN MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 25 | MeONO2 MASS MIXING RATIO AFTER TSTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 27 | C5H8 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 28 | ISOOH MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 29 | ISON MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 30 | MACR MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 31 | MACROOH MASS MIXING RATIO AFTER TS | TDMPMN | DALLTH | UPMEAN |
| 34 | 32 | MPAN MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 33 | HACET MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 34 | MGLY MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 35 | NALD MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 36 | HCOOH MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 37 | MeCO3H MASS MIXING RATIO AFTER TSTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 38 | MeCO2H MASS MIXING RATIO AFTER TSTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 40 | ISO2 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 41 | Cl MASS MIXING RATIO AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 42 | ClO MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 43 | Cl2O2 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 44 | OClO MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 45 | Br MASS MIXING RATIO AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 47 | BrCl MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 48 | BrONO2 MASS MIXING RATIO AFTER TSTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 49 | N2O MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 51 | HOCl MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 52 | HBr MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 53 | HOBr MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 54 | ClONO2 MASS MIXING RATIO AFTER TSTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 55 | CFCl3 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 56 | CF2Cl2 MASS MIXING RATIO AFTER TSTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 57 | MeBr MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 58 | N MASS MIXING RATIO AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 59 | O3P MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 60 | MACRO2 MASS MIXING RATIO AFTER TSTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 70 | H2 MASS MIXING RATIO AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 71 | DMS MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 72 | SO2 MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 73 | H2SO4 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 74 | MSA MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 75 | DMSO MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 76 | NH3 MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 77 | CS2 MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 78 | COS MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 79 | H2S MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 80 | H MASS MIXING RATIO AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 81 | OH MASS MIXING RATIO AFTER TIMESTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 82 | HO2 MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 83 | MeOO MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 84 | EtOO MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 85 | MeCO3 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 86 | n-PrOO MASS MIXING RATIO AFTER TSTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 87 | i-PrOO MASS MIXING RATIO AFTER TSTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 88 | EtCO3 MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 89 | MeCOCH2OO MASS MIXING RATIO AFTER T | TDMPMN | DALLTH | UPMEAN |
| 34 | 90 | MeOH MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 91 | MONOTERPENE MASS MIXING RATIO AFT T | TDMPMN | DALLTH | UPMEAN |
| 34 | 92 | SEC_ORG MASS MIXING RATIO AFTER TS | TDMPMN | DALLTH | UPMEAN |
| 34 | 94 | SO3 MASS MIXING RATIO AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 98 | LUMPED N (as NO2) MMR AFTER TIMESTE | TDMPMN | DALLTH | UPMEAN |
| 34 | 99 | LUMPED Br (as BrO) MMR AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 100 | LUMPED Cl (as HCl) MMR AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 101 | NUCLEATION MODE (SOLUBLE) NUMBER | TDMPMN | DALLTH | UPMEAN |
| 34 | 102 | NUCLEATION MODE (SOLUBLE) H2SO4 MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 103 | AITKEN MODE (SOLUBLE) NUMBER | TDMPMN | DALLTH | UPMEAN |
| 34 | 104 | AITKEN MODE (SOLUBLE) H2SO4 MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 105 | AITKEN MODE (SOLUBLE) BC MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 106 | AITKEN MODE (SOLUBLE) OC MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 107 | ACCUMULATION MODE (SOLUBLE) NUMBER | TDMPMN | DALLTH | UPMEAN |
| 34 | 108 | ACCUMULATION MODE (SOL) H2SO4 MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 109 | ACCUMULATION MODE (SOL) BC MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 110 | ACCUMULATION MODE (SOL) OC MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 111 | ACCUMULATION MODE (SOL) SEA SALT MM | TDMPMN | DALLTH | UPMEAN |
| 34 | 113 | COARSE MODE (SOLUBLE) NUMBER | TDMPMN | DALLTH | UPMEAN |
| 34 | 114 | COARSE MODE (SOLUBLE) H2SO4 MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 115 | COARSE MODE (SOLUBLE) BC MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 116 | COARSE MODE (SOLUBLE) OC MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 117 | COARSE MODE (SOLUBLE) SEA SALT MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 119 | AITKEN MODE (INSOLUBLE) NUMBER | TDMPMN | DALLTH | UPMEAN |
| 34 | 120 | AITKEN MODE (INSOLUBLE) BC MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 121 | AITKEN MODE (INSOLUBLE) OC MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 126 | NUCLEATION MODE (SOLUBLE) OC MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 127 | AITKEN MODE (SOLUBLE) SEA SALT MMR | TDMPMN | DALLTH | UPMEAN |
| 34 | 149 | PASSIVE O3 MASS MIXING RATIO | TDMPMN | DALLTH | UPMEAN |
| 34 | 151 | O(1D) MASS MIXING RATIO | TDMPMN | DALLTH | UPMEAN |
| 34 | 152 | O(3P) MASS MIXING RATIO | TDMPMN | DALLTH | UPMEAN |
| 34 | 154 | BrO MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 155 | HCl MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 156 | Cly MASS MIXING RATIO AFTER TSTEP | TDMPMN | DALLTH | UPMEAN |
| 34 | 157 | AEROSOL SURFACE AREA | TDMPMN | DALLTH | UPMEAN |
| 34 | 158 | NAT PSC MASS MIXING RATIO AFTER TS | TDMPMN | DALLTH | UPMEAN |
| 34 | 159 | OZONE COLUMN | TDAYM | D1TH | UPA |
| 34 | 159 | OZONE COLUMN | TDMPMN | DALLTH | UPMEAN |
| 34 | 160 | Heterogenous Rate for N2O5 | TDMPMN | DALLTH | UPMEAN |
| 34 | 161 | Heterogenous Rates for HO2+HO2 | TDMPMN | DALLTH | UPMEAN |
| 34 | 162 | TOTAL CLOUD DROPLET NO. CONC. (m^-3 | TDMPMN | DALLTH | UPMEAN |
| 34 | 163 | <CLOUD DROPLET NO CONC^(-1/3)> (m) | TDMPMN | DALLTH | UPMEAN |
| 34 | 342 | RXN FLUX: NO3+C5H8->ISON | TMMNUKCA | DALLTH | UPB |
| 34 | 343 | RXN FLUX: NO+ISO2->ISON | TMMNUKCA | DALLTH | UPB |
| 34 | 344 | RXN FLUX: H2O2 PRODUCTION | TMMNUKCA | DALLTH | UPB |
| 34 | 345 | RXN FLUX: ROOH PRODUCTION | TMMNUKCA | DALLTH | UPB |
| 34 | 346 | RXN FLUX: HONO2 PRODUCTION (HNO3) | TMMNUKCA | DALLTH | UPB |
| 34 | 352 | TENDENCY: O3 (TROP ONLY) | TMMNUKCA | DALLTH | UPB |
| 34 | 353 | TROPOSPHERIC O3 | TMNMEAN | DALLTH | UPB |
| 34 | 354 | TENDENCY: O3 (WHOLE ATMOS) | TMMNUKCA | DALLTH | UPB |
| 34 | 363 | AIR MASS DIAGNOSTIC (WHOLE ATMOS) | TDMPMN | DALLTH | UPMEAN |
| 34 | 363 | AIR MASS DIAGNOSTIC (WHOLE ATMOS) | TMNMEAN | DALLTH | UPB |
| 34 | 371 | CO BUDGET: CO LOSS VIA OH+CO | TMMNUKCA | DALLTH | UPB |
| 34 | 372 | CO BUDGET: CO PROD VIA HCHO+OH/NO3 | TMMNUKCA | DALLTH | UPB |
| 34 | 373 | CO BUDGET: CO PROD VIA MGLY+OH/NO3 | TMMNUKCA | DALLTH | UPB |
| 34 | 374 | CO BUDGET: CO MISC PROD&O3+MACR/ISO | TMMNUKCA | DALLTH | UPB |
| 34 | 375 | CO BUDGET: CO PROD VIA HCHO PHOT RA | TMMNUKCA | DALLTH | UPB |
| 34 | 376 | CO BUDGET: CO PROD VIA HCHO PHOT MO | TMMNUKCA | DALLTH | UPB |
| 34 | 377 | CO BUDGET: CO PROD VIA MGLY PHOTOL | TMMNUKCA | DALLTH | UPB |
| 34 | 378 | CO BUDGET: CO PROD VIA MISC PHOTOL | TMMNUKCA | DALLTH | UPB |
| 34 | 379 | CO BUDGET: CO DRY DEPOSITION (3D) | TMMNUKCA | DALLTH | UPB |
| 34 | 381 | LIGHTNING NOx EMISSIONS 3D | TDMPMN | DALLTH | UPMEAN |
| 34 | 382 | LIGHTNING DIAGNOSTIC: TOT FLASHES 2 | TDMPMN | DIAG | UPMEAN |
| 34 | 383 | LIGHTNING DIAG: CLD TO GRND FLSH 2D | TDMPMN | DIAG | UPMEAN |
| 34 | 384 | LIGHTNING DIAG: CLD TO CLD FLSH 2D | TDMPMN | DIAG | UPMEAN |
| 34 | 385 | LIGHTNING DIAG: TOTAL N PRODUCED 2D | TDMPMN | DIAG | UPMEAN |
| 34 | 391 | STRATOSPHERIC OH PRODUCTION | TMMNUKCA | DALLTH | UPB |
| 34 | 392 | STRATOSPHERIC OH LOSS | TMMNUKCA | DALLTH | UPB |
| 34 | 401 | STRAT O3 PROD: O2+PHOTON->O(3P)+O(3 | TMMNUKCA | DALLTH | UPB |
| 34 | 402 | STRAT O3 PROD: O2+PHOTON->O(3P)+O(1 | TMMNUKCA | DALLTH | UPB |
| 34 | 403 | STRAT O3 PROD: HO2 + NO | TMMNUKCA | DALLTH | UPB |
| 34 | 404 | STRAT O3 PROD: MeOO + NO | TMMNUKCA | DALLTH | UPB |
| 34 | 411 | STRAT O3 LOSS: Cl2O2 + PHOTON | TMMNUKCA | DALLTH | UPB |
| 34 | 412 | STRAT O3 LOSS: BrO + ClO | TMMNUKCA | DALLTH | UPB |
| 34 | 413 | STRAT O3 LOSS: HO2 + O3 | TMMNUKCA | DALLTH | UPB |
| 34 | 414 | STRAT O3 LOSS: ClO + HO2 | TMMNUKCA | DALLTH | UPB |
| 34 | 415 | STRAT O3 LOSS: BrO + HO2 | TMMNUKCA | DALLTH | UPB |
| 34 | 416 | STRAT O3 LOSS: O(3P) + ClO | TMMNUKCA | DALLTH | UPB |
| 34 | 417 | STRAT O3 LOSS: O(3P) + NO2 | TMMNUKCA | DALLTH | UPB |
| 34 | 418 | STRAT O3 LOSS: O(3P) + HO2 | TMMNUKCA | DALLTH | UPB |
| 34 | 419 | STRAT O3 LOSS: O3 + H | TMMNUKCA | DALLTH | UPB |
| 34 | 420 | STRAT O3 LOSS: O(3P) + O3 | TMMNUKCA | DALLTH | UPB |
| 34 | 421 | STRAT O3 LOSS: NO3 + PHOTON | TMMNUKCA | DALLTH | UPB |
| 34 | 422 | STRAT O3 LOSS: O(1D) + H2O | TMMNUKCA | DALLTH | UPB |
| 34 | 423 | STRAT O3 LOSS: HO2 + NO3 | TMMNUKCA | DALLTH | UPB |
| 34 | 424 | STRAT O3 LOSS: OH + NO3 | TMMNUKCA | DALLTH | UPB |
| 34 | 425 | STRAT O3 LOSS: NO3 + HCHO | TMMNUKCA | DALLTH | UPB |
| 34 | 431 | STRAT MISC: O3 DRY DEPOSITION (3D) | TMMNUKCA | DALLTH | UPB |
| 34 | 432 | STRAT MISC: NOy DRY DEPOSITION (3D) | TMMNUKCA | DALLTH | UPB |
| 34 | 433 | STRAT MISC: NOy WET DEPOSITION (3D) | TMMNUKCA | DALLTH | UPB |
| 34 | 447 | Monoterp + OH => Sec_org | TMMNUKCA | DALLTH | UPB |
| 34 | 448 | Monoterp + O3 => Sec_org | TMMNUKCA | DALLTH | UPB |
| 34 | 449 | Monoterp + NO3 => Sec_org | TMMNUKCA | DALLTH | UPB |
| 34 | 451 | SO2 + H2O2 => NULL0 | TMMNUKCA | DALLTH | UPB |
| 34 | 452 | SO2 + O3 => NULL1 | TMMNUKCA | DALLTH | UPB |
| 34 | 453 | SO2 + O3 => NULL2 | TMMNUKCA | DALLTH | UPB |
| 34 | 454 | SO2 DRY DEPOSITION (3D) | TMMNUKCA | DALLTH | UPB |
| 34 | 455 | SO2 WET DEPOSITION (3D) | TMMNUKCA | DALLTH | UPB |
| 34 | 460 | CS2+O(3P)->COS+SO2+CO BIMOL | TMMNUKCA | DALLTH | UPB |
| 34 | 461 | CS2+OH->COS+SO2 BIMOL | TMMNUKCA | DALLTH | UPB |
| 34 | 462 | DMS+OH->SO2 BIMOL | TMMNUKCA | DALLTH | UPB |
| 34 | 463 | DMS+OH->MSA+SO2 BIMOL | TMMNUKCA | DALLTH | UPB |
| 34 | 464 | DMS+NO3->SO2 BIMOL | TMMNUKCA | DALLTH | UPB |
| 34 | 465 | DMS+O(3P)->SO2 BIMOL | TMMNUKCA | DALLTH | UPB |
| 34 | 466 | H2S+O(3P)->OH+SO2 BIMOL | TMMNUKCA | DALLTH | UPB |
| 34 | 467 | H2S+OH->SO2+H20 BIMOL | TMMNUKCA | DALLTH | UPB |
| 34 | 468 | COS+O(3P)->CO+SO2 BIMOL | TMMNUKCA | DALLTH | UPB |
| 34 | 469 | COS+OH->COS+SO2 BIMOL | TMMNUKCA | DALLTH | UPB |
| 34 | 470 | SO2+O3->SO3 BIMOL | TMMNUKCA | DALLTH | UPB |
| 34 | 471 | SO3+H2O->H2SO4+H2O BIMOL | TMMNUKCA | DALLTH | UPB |
| 34 | 472 | CS2->COS+SO2 PHOTOL | TMMNUKCA | DALLTH | UPB |
| 34 | 473 | COS->CO+SO2 PHOTOL | TMMNUKCA | DALLTH | UPB |
| 34 | 474 | H2SO4->SO3+OH PHOTOL | TMMNUKCA | DALLTH | UPB |
| 34 | 475 | SO3->SO2+O(3P) PHOTOL | TMMNUKCA | DALLTH | UPB |
| 34 | 476 | SO2+OH->SO3+HO2 | TMMNUKCA | DALLTH | UPB |
| 34 | 477 | DMSO DRY DEP | TMMNUKCA | DALLTH | UPB |
| 34 | 478 | NH3 DRY DEP | TMMNUKCA | DALLTH | UPB |
| 34 | 479 | Monoterp DRY DEP | TMMNUKCA | DALLTH | UPB |
| 34 | 480 | SEC_ORG DRY DEP | TMMNUKCA | DALLTH | UPB |
| 34 | 481 | O3 WET DEP | TMMNUKCA | DALLTH | UPB |
| 34 | 482 | DMSO WET DEP | TMMNUKCA | DALLTH | UPB |
| 34 | 483 | NH3 WET DEP | TMMNUKCA | DALLTH | UPB |
| 34 | 484 | SEC_ORG WET DEP | TMMNUKCA | DALLTH | UPB |
| 35 | 1 | O3 MASS MIX RATIO ON PRESS LEVELS | TDAYM | DP36CCM | UPA |
| 35 | 1 | O3 MASS MIX RATIO ON PRESS LEVELS | TDMPMN | DP36CCM | UPMEAN |
| 35 | 173 | HEAVYSIDE FUNC FOR P LEVEL DIAGS | TDAYM | DP36CCM | UPA |
| 35 | 173 | HEAVYSIDE FUNC FOR P LEVEL DIAGS | TDMPMN | DP36CCM | UPMEAN |
| 35 | 341 | FLUX: OH+CH4 REACTION ON PLEVS | TDAYM | DP36CCM | UPA |
| 35 | 341 | FLUX: OH+CH4 REACTION ON PLEVS | TDMPMN | DP36CCM | UPMEAN |
| 38 | 201 | PRIMARY SO4 TO AITKEN (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 202 | PRIMARY SO4 TO ACCUMULATION (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 203 | PRIMARY SO4 TO COARSE (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 204 | PRIMARY SEA-SALT TO ACCUMULTN (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 205 | PRIMARY SEA-SALT TO COARSE (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 207 | PRIMARY BLACK CARBON TO AITKEN (INS | TMMNUKCA | DALLTH | UPB |
| 38 | 209 | PRIMARY ORG. CARBON TO AITKEN (INS | TMMNUKCA | DALLTH | UPB |
| 38 | 214 | DRY DEPOSITION SO4 NUCLEATION (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 215 | DRY DEPOSITION SO4 AITKEN (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 216 | DRY DEPOSITION SO4 ACCUMULTN (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 217 | DRY DEPOSITION SO4 COARSE (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 218 | DRY DEPOSITION SEA-SALT ACC (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 219 | DRY DEPOSITION SEA-SALT COARSE (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 220 | DRY DEPOSITION BLK CARB AITKEN (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 221 | DRY DEPOSITION BLK CARB ACCUM. (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 222 | DRY DEPOSITION BLK CARB COARSE (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 223 | DRY DEPOSITION BLK CARB AITKEN (INS | TMMNUKCA | DALLTH | UPB |
| 38 | 224 | DRY DEPOSITION ORG CARB NUCLN. (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 225 | DRY DEPOSITION ORG CARB AITKEN (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 226 | DRY DEPOSITION ORG CARB ACCUM. (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 227 | DRY DEPOSITION ORG CARB COARSE (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 228 | DRY DEPOSITION ORG CARB AITKEN (INS | TMMNUKCA | DALLTH | UPB |
| 38 | 239 | NUC SCAVENGING SO4 ACCUMULTN (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 240 | NUC SCAVENGING SO4 COARSE (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 241 | NUC SCAVENGING SEA-SALT ACCUM. (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 242 | NUC SCAVENGING SEA-SALT COARSE (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 243 | NUC SCAVENGING BLK CARB AITKEN (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 244 | NUC SCAVENGING BLK CARB ACCUM. (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 245 | NUC SCAVENGING BLK CARB COARSE (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 246 | NUC SCAVENGING BLK CARB AITKEN (INS | TMMNUKCA | DALLTH | UPB |
| 38 | 247 | NUC SCAVENGING ORG CARB NUCLN. (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 248 | NUC SCAVENGING ORG CARB AITKEN (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 249 | NUC SCAVENGING ORG CARB ACCUM. (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 250 | NUC SCAVENGING ORG CARB COARSE (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 251 | NUC SCAVENGING ORG CARB AITKEN (INS | TMMNUKCA | DALLTH | UPB |
| 38 | 261 | IMP SCAVENGING SO4 NUCLN. (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 262 | IMP SCAVENGING SO4 AITKEN (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 263 | IMP SCAVENGING SO4 ACCUMULTN (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 264 | IMP SCAVENGING SO4 COARSE (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 265 | IMP SCAVENGING SEA-SALT ACCUM. (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 266 | IMP SCAVENGING SEA-SALT COARSE (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 267 | IMP SCAVENGING BLK CARB AITKEN (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 268 | IMP SCAVENGING BLK CARB ACCUM. (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 269 | IMP SCAVENGING BLK CARB COARSE (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 270 | IMP SCAVENGING BLK CARB AITKEN (INS | TMMNUKCA | DALLTH | UPB |
| 38 | 271 | IMP SCAVENGING ORG CARB NUCLN. (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 272 | IMP SCAVENGING ORG CARB AITKEN (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 273 | IMP SCAVENGING ORG CARB ACCUM. (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 274 | IMP SCAVENGING ORG CARB COARSE (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 275 | IMP SCAVENGING ORG CARB AITKEN (INS | TMMNUKCA | DALLTH | UPB |
| 38 | 285 | INCLOUD SO4 (H2O2) TO ACCUMLTN (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 286 | INCLOUD SO4 (H2O2) TO COARSE (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 288 | INCLOUD SO4 (O3) TO ACCUMULTN (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 289 | INCLOUD SO4 (O3) TO COARSE (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 290 | CLOUDPROC SU AITSOL --> ACCSOL | TMMNUKCA | DALLTH | UPB |
| 38 | 291 | CLOUDPROC BC AITSOL --> ACCSOL | TMMNUKCA | DALLTH | UPB |
| 38 | 292 | CLOUDPROC OC AITSOL --> ACCSOL | TMMNUKCA | DALLTH | UPB |
| 38 | 294 | CONDENSATION SO4 TO NUCLEATION (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 295 | CONDENSATION SO4 TO AITKEN (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 296 | CONDENSATION SO4 TO ACCUMULTN (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 297 | CONDENSATION SO4 TO COARSE (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 298 | CONDENSATION SO4 TO AITKEN (INSOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 301 | CONDENSATION OC1 TO NUCLEATION (SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 302 | CONDENSATION OC1 TO AITKEN (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 303 | CONDENSATION OC1 TO ACCUMULTN (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 304 | CONDENSATION OC1 TO COARSE (SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 305 | CONDENSATION OC1 TO AITKEN (INSOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 319 | NUCLEATION OF SO4 (TO NUC-SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 320 | COAG SO4 NUCLTN-SOL TO AITKEN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 321 | COAG SO4 NUCLTN-SOL TO ACCUMULN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 322 | COAG SO4 NUCLTN-SOL TO COARSE-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 323 | COAG SO4 NUCLTN-SOL TO AITKEN-INSOL | TMMNUKCA | DALLTH | UPB |
| 38 | 326 | COAG OC1 NUCLTN-SOL TO AITKEN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 327 | COAG OC1 NUCLTN-SOL TO ACCUMULN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 328 | COAG OC1 NUCLTN-SOL TO COARSE-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 329 | COAG OC1 NUCLTN-SOL TO AITKEN-INSOL | TMMNUKCA | DALLTH | UPB |
| 38 | 338 | COAG SO4 AITKEN-SOL TO ACCUMULN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 339 | COAG SO4 AITKEN-SOL TO COARSE-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 342 | COAG BC AITKEN-SOL TO ACCUMULN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 343 | COAG BC AITKEN-SOL TO COARSE-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 346 | COAG OC1 AITKEN-SOL TO ACCUMULN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 347 | COAG OC1 AITKEN-SOL TO COARSE-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 354 | COAG SO4 ACCUMULN-SOL TO COARSE-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 356 | COAG BC ACCUMULN-SOL TO COARSE-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 358 | COAG OC1 ACCUMULN-SOL TO COARSE-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 360 | COAG SEA-SALT ACC-SOL TO COARSE-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 366 | COAG BC AITKEN-INS TO ACCUMULN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 367 | COAG OC1 AITKEN-INS TO ACCUMULN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 368 | COAG BC AITKEN-INS TO COARSE-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 371 | AGEING SO4 AITKEN-INS TO AITKEN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 372 | AGEING BC AITKEN-INS TO AITKEN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 373 | AGEING OC1 AITKEN-INS TO AITKEN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 375 | MERGE SO4 NUCLTN-SOL TO AITKEN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 376 | MERGE OC1 NUCLTN-SOL TO AITKEN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 378 | MERGE SO4 AITKEN-SOL TO ACCUMULN-SO | TMMNUKCA | DALLTH | UPB |
| 38 | 379 | MERGE BC AITKEN-SOL TO ACCUMULN-SO | TMMNUKCA | DALLTH | UPB |
| 38 | 380 | MERGE OC1 AITKEN-SOL TO ACCUMULN-SO | TMMNUKCA | DALLTH | UPB |
| 38 | 382 | MERGE SO4 ACCUMULN-SOL TO COARSE-SO | TMMNUKCA | DALLTH | UPB |
| 38 | 383 | MERGE SEA_SALT ACC-SOL TO COARSE-SO | TMMNUKCA | DALLTH | UPB |
| 38 | 384 | MERGE BC ACCUMULN-SOL TO COARSE-SO | TMMNUKCA | DALLTH | UPB |
| 38 | 385 | MERGE OC1 ACCUMULN-SOL TO COARSE-SO | TMMNUKCA | DALLTH | UPB |
| 38 | 401 | DRY PARTICLE DIAMETER NUCLEATION-SO | TMMNUKCA | DALLTH | UPB |
| 38 | 402 | DRY PARTICLE DIAMETER AITKEN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 403 | DRY PARTICLE DIAMETER ACCUMULATN-SO | TMMNUKCA | DALLTH | UPB |
| 38 | 404 | DRY PARTICLE DIAMETER COARSE-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 405 | DRY PARTICLE DIAMETER AITKEN-INS | TMMNUKCA | DALLTH | UPB |
| 38 | 408 | WET PARTICLE DIAMETER NUCLEATION-SO | TMMNUKCA | DALLTH | UPB |
| 38 | 409 | WET PARTICLE DIAMETER AITKEN-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 410 | WET PARTICLE DIAMETER ACCUMULATN-SO | TMMNUKCA | DALLTH | UPB |
| 38 | 411 | WET PARTICLE DIAMETER COARSE-SOL | TMMNUKCA | DALLTH | UPB |
| 38 | 412 | WATER CONTENT FOR NUCLEATION-SOLUBL | TMMNUKCA | DALLTH | UPB |
| 38 | 413 | WATER CONTENT FOR AITKEN -SOLUBL | TMMNUKCA | DALLTH | UPB |
| 38 | 414 | WATER CONTENT FOR ACCUMULTN.-SOLUBL | TMMNUKCA | DALLTH | UPB |
| 38 | 415 | WATER CONTENT FOR COARSE -SOLUBL | TMMNUKCA | DALLTH | UPB |
| 38 | 437 | CN NUMBER CONCENTRATION | TMMNUKCA | DALLTH | UPB |
| 38 | 438 | CCN NO. CONCENTRN. (ACC+COR) | TMMNUKCA | DALLTH | UPB |
| 38 | 439 | CCN NO. CONCENTRN. (ACC+COR+AIT>25r | TMMNUKCA | DALLTH | UPB |
| 38 | 440 | CCN NO. CONCENTRN. (ACC+COR+AIT>35r | TMMNUKCA | DALLTH | UPB |
| 38 | 441 | CD NUMBER CONCENTRATION | TMMNUKCA | DALLTH | UPB |
| 38 | 469 | ACTIVE CLOUD DROP NO CONC (NUC-SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 470 | ACTIVE CLOUD DROP NO CONC (AIT-SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 471 | ACTIVE CLOUD DROP NO CONC (ACC-SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 472 | ACTIVE CLOUD DROP NO CONC (COR-SOL) | TMMNUKCA | DALLTH | UPB |
| 38 | 476 | CLOUD LIQUID FRACTION | TMMNUKCA | DALLTH | UPB |
| 38 | 477 | TOTAL CDNC * CLOUD LIQ FRACTION | TMMNUKCA | DALLTH | UPB |
ARCHER Required Changes
On ARCHER (job xkawe), the EP flux diagnostics (s30i310-316) do not work, either in the climate meaning stream or another pp-stream.
When using these with UPMEAN the ATP traceback for this error is
Application 9859855 is crashing. ATP analysis proceeding... ATP Stack walkback for Rank 0 starting: _start@start.S:113 __libc_start_main@libc-start.c:226 flumemain_@flumeMain.f90:48 um_shell_@um_shell.f90:1865 u_model_@u_model.f90:3730 meanctl_@meanctl.f90:3631 acumps_@acumps.f90:1475 general_scatter_field_@general_scatter_field.f90:1098 stash_scatter_field_@stash_scatter_field.f90:955 gcg_ralltoalle_@gcg_ralltoalle.f90:180 gcg__ralltoalle_multi_@gcg_ralltoalle_multi.f90:335 ATP Stack walkback for Rank 0 done Process died with signal 11: 'Segmentation fault' Forcing core dumps of ranks 0, 1, 12, 26, 118
When moving these to e.g. UPB and TMONMEAN, the model completes the NRUN stage (currently 3 months) but then hangs in the CRUN step. I have not found a fix for this.
For the ARCHER run, some diagnostics were moved to UPB from UPMEAN. This means that while monthly means were produced, seasonal and annual means were not.
I have raised a ticket with the NCAS-CMS helpdesk on this issue.
Contributions and Acknowledgements
Luke Abraham would like to thank the following people (in no particular order) for their help in creating this job:
- Mohit Dalvi, Met Office
- Alex Archibald, NCAS/University of Cambridge
- Graham Mann, NCAS/University of Leeds
- Colin Johnson, Met Office
- Fiona O'Connor, Met Office
- Nick Savage, Met Office
- Sandip Dhomse, University of Leeds
- Martyn Chipperfield, NCAS/University of Leeds
- James Keeble, University of Cambridge
- Paul Telford, NCAS/University of Cambridge
- Maria Russo, NCAS/UNiversity of Cambridge
- John Pyle, NCAS/University of Cambridge
- Zak Kipling, University of Oxford
- Rosalind West, University of Oxford
- Philip Stier, University of Oxford
- Karthee Sivalingam, NCAS/University of Reading
- Kirsty Pringle, University of Leeds
- Ken Carslaw, NCAS/University of Leeds
- Ben Johnson, Met Office
- Jane Mulcahy, Met Office
- Nicolas Bellouin, University of Reading
- Peter Uhe, CSIRO
- James Mollard, University of Reading
- NCAS-CMS and the Met Office MONSooN team