UKCA Chemistry and Aerosol vn10.4 Tutorial 11
UKCA Chemistry and Aerosol Tutorials at vn10.4
Back to UKCA Chemistry and Aerosol Tutorials
What you will learn in this Tutorial
In this tutorial you will learn about the GLOMAP-mode aerosol module and how it tracks different aerosol types within several size classes. You will understand the standard configuration used in the UKCA jobs so far whereby the mass mixing ratios of sulphate, sea-salt, black carbon and organic matter in each mode are transported via separate tracers. GLOMAP-mode is an aerosol microphysics scheme and therefore, as well as transporting the mass of several components in the modes, the scheme also transports the number concentrations of particles in each mode.
Task 10 already introduced the basic concepts behind the GLOMAP-mode aerosol microphysics scheme and how it differs from the mass-based CLASSIC scheme which preceded UKCA.
Initially developed in the TOMCAT CTM environment (see Manktelow et al., 2007; Mann et al., 2010; Mann et al., 2012), the GLOMAP code then became the aerosol module for the UKCA sub-model of the UM (see Bellouin et al., 2013; Kipling et al., 2013; West et al., 2014; Mann et al., 2014; Dhomse et al., 2014; Turnock et al., 2015; Turnock et al., 2016; Zanchettin et al., 2016).
GLOMAP is now also implemented into the ECMWF Integrated Forecasting System as part of the "Composition IFS" module (C-IFS) where it will be used in combination with data assimilation of satellite Aerosol Optical Depth to provide forecasts and re-analyses of atmospheric composition and boundary conditions for regional air quality models. The shift from the current operational aerosol scheme in C-IFS over to GLOMAP-mode is scheduled to take place during 2018.
The GLOMAP-mode code allows several alternative "aerosol configurations" to be run using the same set of FORTRAN subroutines.
In section 12 of the UKCA UMDP, Table 18 shows the standard configuration for GLOMAP in both of these other 2 host model frameworks (TOMCAT and C-IFS-GLOMAP).
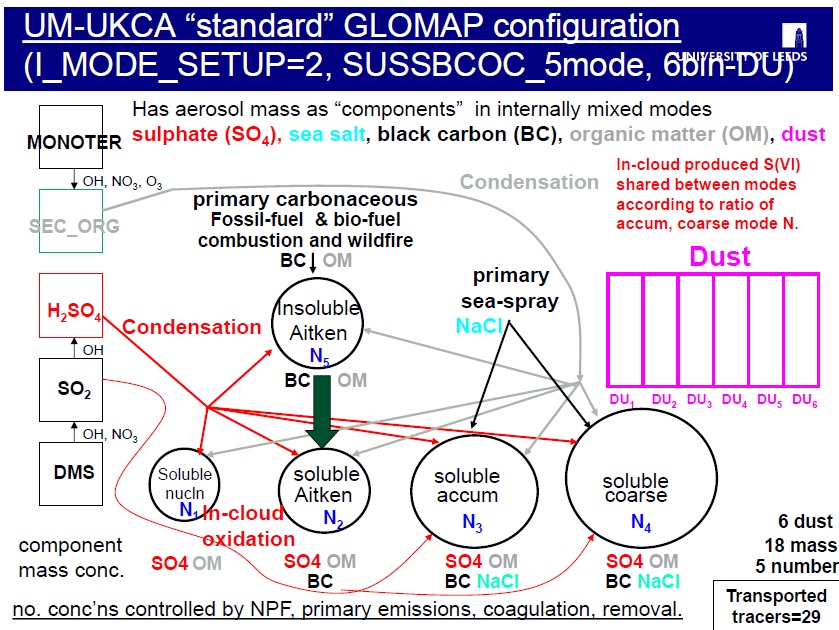
In this 7-mode configuration (known as setup 8), GLOMAP runs with all 7 modes activated, with 2 being pure dust modes and the other 5 containing mixtures of different aerosol components (sulphate, black carbon, organic matter, sea-salt and dust).
With this GLOMAP setup 8, the model runs has 7 number mixing ratios (one for each mode) and has a total of 19 component mass mixing ratios over all the different types.
As mentioned in task 10 however, when GLOMAP is run within UM-UKCA, dust is usually handled by the existing 6-bin UM scheme, and GLOMAP is configured to use the "5-mode configuration" (known as setup 2) covering only 4 of the above 5 components (sulphate, black carbon, organic matter and sea-salt).
The scheme can also be further reduced to cover just the sulphate and sea-salt components in 4 modes (known as setup 1), which was the initial configuration used for interactive stratospheric aerosol simulations (e.g. Dhomse et al., 2014).
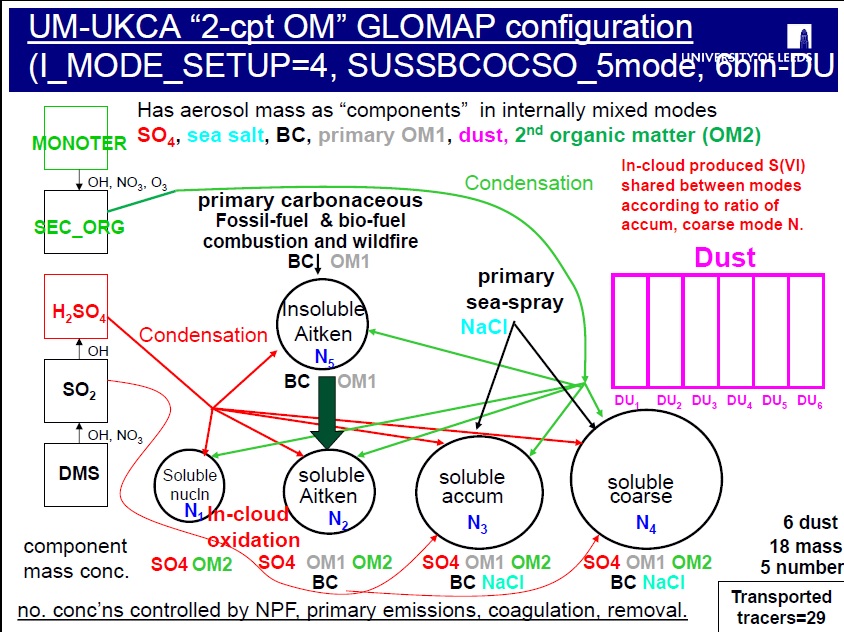
The GLOMAP scheme has been run in the TOMCAT CTM in a range of configurations, including one specifically designed to track two separate components for organic matter (OM), with the I_MODE_SETUP=4 configuration designed to have one component track the primary OM in each mode and a separate OM component to track secondary organic matter in each mode.
Section 12.2 of the UMDP has a more detailed explaination of these configurations with Table 19 showing how these 4 different GLOMAP-mode setups map onto the model tracers.
In this task you will take a copy of the standard UKCA job (which uses GLOMAP-mode setup 2, MS2) and change it to use GLOMAP-mode setup 4 (MS4) to track two separate organic matter (OM) components rather than the usual 1. With the 2-component OC configuration, the model tracks primary (emitted) organic carbon in the usual OM component and secondary organic matter (formed following oxidation in the atmosphere) separately in a 2nd OM component.
Task 11.1: Understand how the GLOMAP aerosol module tracks aerosol species and modes
TASK 11.1: Read section 12 (page 32) of the v8.4 UM Documentation Paper and refer to Tables 18, 19 and 20 on pages 33, 34 and 35.
Task 11.2: Run a copy of the standard UKCA job which tracks two OM components in the GLOMAP modes
Take a copy of the suite you generated from Task 10 with the GLOMAP AOD diagnostics added. This was your copy of the standard ARCHER v10.4 UKCA tutorial suite (u-ai071). The task is to change the aerosol settings from the standard GLOMAP aerosol configuration (setup 2) to instead use the 2-component OM configuration (setup 4).
To run the 2-component GLOMAP configuration, you will need to add in an additional branch to the suite (which supports that) and also make change to the Rose GUI metadata for the suite, so that an extra option for I_MODE_SETUP=4 is made available within the "GLOMAP configuration" selection-button.
First, add in the extra branch. Go to the fcm_make app (4th from the top) and then click on "env" then "Sources" which then shows you the 4 branches that are already used in the UKCA tutorial job. These 4 branches are to enable additional settings over and above what was present in the UM trunk code at v10.4.
To add in the extra branch click the plus sign and then paste the following into the new entry that appears after you click the plus:
branches/dev/grahammann/vn10.4_support_GLOMAPsetup4_for_2cptOM@32212
This is picking up the code-changes from revision 32212 of the branch I put together to enable UM-UKCA to correctly allign with the code that is present for GLOMAP setup 4 (and adds a few bits that were missing in the v10.4 trunk).
Next we'll need to change the GLOMAP setup. In the Rose GUI, you need to open the UM app (click on the right-hand-pointing triangle at the bottom next to "um") and then similarly click on "namelist" then "UM Science settings" and finally you can then select "Section 34: UKCA" to view which options have been selected in this particular UKCA tutorial job.
Scrolling down you will eventually see the UKCA aerosol settings and you'll see "i_mode_setup" with the description "Set aerosol species and modes". In the aerosol configuration used by the job you can see I_MODE_SETUP has been set to 2. Which corresponds to the "standard GLOMAP setup" explained and illustrated above.
You see also that there are alternative options there that can change the aerosol setup to 1 (sulphate and sea-salt in 4 modes), 6 (dust-only in 2 modes) and 8 (sulphate, sea-salt, BC, POM and dust in 7 modes).
In our case, we actually want a different set-up though to any of these standard 4 options -- to activate to use the 2-component organic matter configuration. Of course one needs to know that that configuration is actually available in the code, and although this is explained in the UMDP, it turns out that, at this version, the I_MODE_SETUP=1, 6 and 8 configurations are not supported, and we recommend you do not select those options without corresponding closely with the GLOMAP development team at Leeds.
In order to do this requires to point the Rose GUI to an updated version of the metadata that provides the information for the different panels in the GUI. At the moment the metadata used in the job tells that UM that the only options available are I_MODE_SETUP=1, 2, 6 and 8. But with the changes implemented in the branch above, it will then be possible to also run the model with I_MODE_SETUP=4 (which is the GLOMAP configuration that we want to run with).
The metadata for the UM app within the Rose GUI panel is specified in the first panel selections in the GUI. Instead of clicking on the right-hand-pointing triangle, click on the word um and then you should see a panel with only one entry that says "meta" with the text set as:
/home/grenville/meta/ga7_vn10.4
This is pointing to a rose-meta.conf file that specifies the metadata for the UM app. It is off Grenville Lister's directory because there were things that were decided were required to be added (chosen according to some priorities or requirements) to the functionality of that UM app at that particular time.
In preparation for this task I have update those Rose selections to additionally allow the user to select the option 4 as well as those 1, 2, 6 and 8.
If you change the text set in that box to instead be set as:
/home/gmann/meta/ga7_vn10.4_MS4updated
it will then know that that particular option is available, and the GUI will display it (whereas it didn't with the above path specified).
To get an idea of what is different about the rose-meta.conf file, that then allows the user to choose that option, you could use a graphical difference engine like "tkdiff" or "xxdiff" to compare the rose-meta.conf in those two directories.
tkdiff /home/grenville/meta/ga7_vn10.4/rose-meta.conf /home/gmann/meta/ga7_vn10.4_MS4updated/rose-meta.conf
By doing this, you see that the functionality that was added in the file off Grenville Lister's directory was to enable a new switch to allow the user to scale up or down how much NOx is emitted from lightning sources.
In the hand-edit for the MS4updated version of this, the code has actually provided a replacement for the existing metadata that enabled that switch for different values of I_MODE_SETUP which now allows to take 5 alternative values (1, 2, 4, 6 and 8) instead of the existing 4.
Once you have activated the new meta data you may not be able to seee it until you have saved and exit the GUI and opened it up again.
But once it's refreshed it should be fine to go ahead and select that I_MODE_SETUP=4 option rather than the previous (default) setting of I_MODE_SETUP=2.
When these buttons are selected, the code itself does several things automatically to only activate those tracers required to be switched on.
At previous versions of the UM, one needed to set several hand-edits to edits the file SIZES setting the values of the array TC_UKCA which specifies which of the UKCA tracers are switched on (=1) or off (=0). But at v10.4 this is done automatically within the code itself.
Still it is worth seeing in the code where this is done and I recommend at this point that you do a "checkout" to your local space on PUMA of the source code behind the v10.4 branch that was added above.
Look in the directory src/atmosphere/UKCA for the routine ukca_init.F90 and this gives you some idea of how the code proceesses these different aerosol configurations and how the GLOMAP aerosol tracers in general are specified in the code via the array nm_spec.
In the standard tracer configuration for UKCA specified in this config_plume_scav_on_st.ed hand-edit, tracers Ait_SOL_OC (index 106), Acc_SOL_OC (110), Cor_SOL_OC (116), Ait_INS_OC (121) and Nuc_SOL_OC (126) are set to 1 but tracers Nuc_SOL_SO (128), Ait_SOL_SO (129), Acc_SOL_SO (130) and Cor_SOL_SO (131) are set to zero.
To run with the 2-component OC configuration, the additional "SO" tracers to store the 2nd organic matter component in each mode need to be switched on.
You see that running with the standard configuration of GLOMAP (I_MODE_SETUP=2) requires 83 tracers in the UKCA CheST configuration, with 20 coming from GLOMAP.
To run with the 2-component OM configuration of GLOMAP (I_MODE_SETUP=4) and the UKCA CheST chemistry required 3 addititional aerosol tracers, giving 86 in total.
The extra tracers for the OC2 mass mixing ratios in each are stored in STASH items 128, 129, 130 and 131 in section 34. You see that these are set to 1 for the SO components in each of the soluble modes and the nucleation soluble OC mmr is no longer required as it has been replaced with SO mmr.
In your job add extra daily-mean STASH requests for these additional 4 OM mmr's (as you did to request the standard OM mmrs in task 10).
Also, whereas at v8.4 the user had to switch to a different version of the RADAER hand-edit raderv2_vn84_ARCHER.ed to allow the 2-component OM configuration of GLOMAP to couple to the UM radiation scheme, at v10.4 this is all taken care of automatically.
So the AOD's calculated by RADAER will include the partial volume from each of the OC2 mmr's (items 128 to 131, which are now tracking the secondary OM) as well as the usual OC mmrs (which now only include the primary OM).
Finally, since you have asked the model to run with additional tracers, you also need to specify how these should be initialised.
Go to the Initialisation of User Prognostics panel off the STASH window. Scroll down until you see items 34128, 34129, 34130, 34131 and set the Option column to 3 so that these tracers are initialised to zero values for an NRUN.
Worked Solution
The worked solution to this task can be found in job xjrnm.
Sample output can be found on ARCHER in the directory
/work/n02/n02/ukca/Tutorial/vn8.4/sample_output/Task11.2
Task 11.3 Examine the simulated total organic carbon in the original and two-cpt OC configurations
In the above Task 11.2 you ran a 2-component OC version of the UKCA tutorial
job (xjrnk).
You can also refer to the worked solution xjrnm which I have
configured in this way.
See that xjrnl was the same as the UKCA tutorial job
xjrnk except that I have added the extra STASH requests as
in Task 10.3.
So by now you should have equivalent standard (as xjrnl) and
2-component OM (as xjrnm) versions of the UKCA tutorial job.
In these jobs you have requested numerous daily-mean fields to be output in
the .pa files.
So in your /work/n02/n02/ directory on ARCHER you should have jobida.pa19991201 files for your standard and 2-component OM jobs.
Included in the extra STASH requests are the mass mixing ratios of OC (the standard organic component) and SO (the 2nd organic component) in each mode.
The OC mmrs are STASH section 34, items 126 (nucleation mode), 106 (Aitken-soluble), 110 (accumn-soluble), 116 (coarse-soluble) and 121 (Aitken-insoluble).
The SO mmrs are STASH section 34, items 128 (nucleation mode), 129 (Aitken-soluble), 130 (accumn-soluble), 131 (coarse-soluble).
These STASH item numbers and the details of the standard and 2-component GLOMAP configurations can be found in the UKCA UMDP section 12 Tables 19 and 20.
Note that there is no SO in the Aitken-insoluble mode as this contains only primary carbonaceous particles. Any SO or OC condensing onto the particles in the insoluble modes is immediately transferred over to the corresponding soluble mode following the "condensation-ageing" approach used by the model. This OC or SO condensing onto the insoluble particles is a kind of "coating" for the particles making the particles hygroscopic/soluble.
You could also try adding STASH requests for the mmr of the gas phase species MONOTER and SEC_ORG (STASH section 34, items 91 and 92).
As an example I have put here a link to a pdf ![]() OMcomparison
OMcomparison ![]() showing global maps comparing surface OM fields between the worked solutions
showing global maps comparing surface OM fields between the worked solutions
xjrnl (top-left, labelled as xkwhg) and
xjrnm (top-right, labelled as xkwhh).
Page 1 of the pdf compares the "total POM mmr" at the surface which is the total particulate organic matter (POM) summing up the mass of OC and SO in each mode.
Pages 2 and 3 show comparisons of "total POM1 mmr" and "total POM2 mmr" which are the sum of the 1st and 2nd organic component over all the modes.
You can see from the example that the "total POM2 mmr" in xjrnl
is zero everywhere. That's because in this job GLOMAP has the standard configuration
with just one organic component.
By contrast the "total POM2 mmr" for xjrnm has considerable
concentrations in vegetated continental regions.
In this "I_MODE_SETUP=4" configuration, the "SEC_ORG" species (which contains the
secondary organics from monoterpene oxidation) condenses into the "SO" component,
whereas in xjrnl SEC_ORG condenses into the "OC" component.
The bottom left panel on each page shows a global map of the ratio of the field for the two model runs. One can use this kind of approach to track the fraction of the OM that is biogenic and anthropogenic.
Note however that we initialised the SO mmr's to zero at the start of the 1-day run. So the OC1 mmrs will be spinning down and the SO mmrs will be spinning up. The daily-mean values are averaging over values on each the 1-hour timesteps over which the UKCA chemistry and aerosol processes are integrated. So although the ratio shown in the bottom-left on page 2 is indicative of the biogenic fraction it should be treated with caution as the fields will not have spun-up/down yet.
This task illustrates how one can separate out the aerosol mass from different sources and track them separately via a different aerosol component.
One could also introduce a 2nd gas phase species like "SEC_ORG" to track different types of SOA. For example one could configure the model so that such a 2nd "SEC_ORG2" species held semi-volatile oxidised organic species with only the very low volatility oxidised organics held in the usual "SEC_ORG" species.
Written by Graham Mann 2016