UKCA Chemistry and Aerosol vn10.4 Tutorial 12
UKCA Chemistry and Aerosol Tutorials at vn10.4
What you will learn in this Tutorial
In this tutorial you will learn about how to quantify the radiative effects of aerosol simulated by GLOMAP-mode in UM-UKCA.
In the first task you will update your copy of the UKCA tutorial job to request radiative fluxes allowing the radiative flux perturbation (or effective radiative forcing) to be diagnosed based on difference in the fluxes between a pair of UM-UKCA jobs with some difference (e.g. pre-industrial and present-day emissions jobs).
The second task involves configuring a copy of the UKCA tutorial job to run in double-call configuration whereby the aerosol radiative effects can be diagnosed at each radiation timestep.
Task 12.1: Update your copy of the UKCA tutorial job to diagnose Top Of the Atmosphere (TOA) radiative fluxes
In this task you will add STASH requests for SW and LW outgoing radiative fluxes at the top of the atmosphere to enable the radiative forcing from a particular change to be diagnosed.
The user should note however that to illustrate the task we are adding these requests to the UKCA tutorial job which is just a 1-day simulation.
One would need to average the flux-difference between the pair of simulations over an appropriate timescale (e.g. multi-annual monthly-means) in order to diagnose an effective radiative forcing appropriately.
Noting the above caveat, proceed and add daily-mean STASH requests for section 1 item 208 (all-sky outgoing short wave flux at the top-of-the-atmosphere) and section 2 item 205 (all-sky outgoing long wave flux at the top-of-the-atmosphere) to the updated copy of the UKCA tutorial suite you produced in Task 10 after adding AOD and OM diags (the reference job for this is u-ai830).
The radiative fluxes are 2-dimensional diagnostics (longitude by latitude) so you should use the DIAG domain profile in this case. For daily-means use the TDAYM time profile. Again, since we require the daily-mean fluxes to be output to the .pa file you should request the diagnostics with the UPK usage profile.
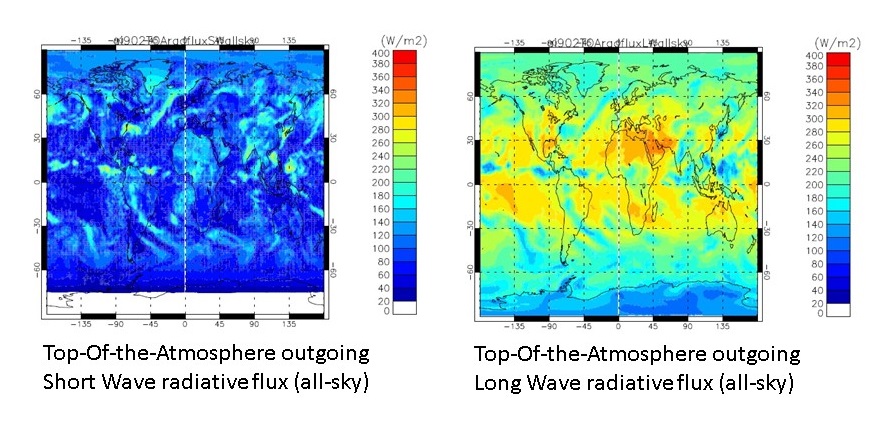
The Figure below shows the daily-mean SW and LW all-sky TOA radiative flux fields from the UKCA tutorial job for 1st September 1999 (produced by gmann job u-ai902, which updated from u-ai830 by adding in the requests).
Example Output
You .pk file should now contain the following fields:
0 : 192 144 1 1 Upward solar 1 : 192 144 1 1 IR up 2 : 192 144 6 1 Unspecified 3 : 192 144 6 1 Unspecified 4 : 192 144 6 1 Unspecified 5 : 192 144 6 1 Unspecified 6 : 192 144 6 1 Unspecified 7 : 192 144 6 1 Unspecified 8 : 192 144 6 1 Unspecified 9 : 192 144 6 1 Unspecified 10 : 192 144 6 1 Unspecified 11 : 192 144 6 1 Unspecified 12 : 192 144 6 1 Unspecified 13 : 192 144 6 1 Unspecified 14 : 192 144 85 1 AITKEN MODE (SOLUBLE) OC MMR 15 : 192 144 85 1 ACCUMULATION MODE (SOL) OC MMR 16 : 192 144 85 1 COARSE MODE (SOLUBLE) OC MMR 17 : 192 144 85 1 AITKEN MODE (INSOLUBLE) OC MMR 18 : 192 144 85 1 NUCLEATION MODE (SOLUBLE) OC MMR
Example output for Task12.1 can be found on ARCHER in the following directory:
/work/n02/n02/ukca/Tutorial/vn10.4/sample_output/Task12.1
The Rose specific changes required are:
Index: app/um/rose-app.conf =================================================================== --- app/um/rose-app.conf (revision 28624) +++ app/um/rose-app.conf (revision 28651) @@ -6638,6 +6638,14 @@ tim_name='TRADDM' use_name='UPMEAN' +[namelist:streq(678928ac)] +dom_name='DIAG' +isec=1 +item=208 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(67a9602e)] dom_name='DIAGAOT' isec=2 @@ -8878,6 +8886,14 @@ tim_name='TDAYM' use_name='UPJ' +[namelist:streq(c8bb6a1f)] +dom_name='DIAG' +isec=2 +item=205 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(c8c613fb)] dom_name='DALLTH' isec=34
A file containing these changes can be found at:
/home/ukca/Tutorial/worked_solutions/Task12.1/task12.1.rose.diff
Task 12.2 Configure the UKCA tutorial job to run as a double-call job diagnosing aerosol radiative effects
In this task you will copy the copy of the standard tutorial job you used in Task 12.1
(u-ai902)
and configure it to run with double-call to the radiation scheme to diagnose the radiative
effects of the aerosol simulated by GLOMAP in UM-UKCA.
The UM has been coded to allow the user to diagnose radiative effects of a particular forcing agent by calling the radiation scheme twice with one of the calls setting the agent's concentration to zero. Special forcing STASH items are included within the UM which store the difference in the radiative fluxes between the two radiation calls.
In the um app in the Rose GUI go to namelist --> UM Science Settings and then choose Section 1 - 2: Radiation.
In the General Options panel, you see that at the top there is an i_rad_extra_call "Options for multiple calls to radiation" button-selector.
The UKCA tutorial job is set to "Single call" which is the standard way of running the UM with a single call to the radiation scheme every radiation timestep (here one hour). You see there is a 2nd option there to select "Diagnose radiative forcings". Selecting this option activates the double-call approach where the radiation scheme is called twice on each radiation timestep once with the forcing agent and once without it.
With this option, the idea is that the user can then isolate the difference in the radiative fluxes (i.e. the forcing) due to a particular forcing agent of interest. There are a host of additional settings that become available when you select this option setting exactly which forcings to isolate via the difference between the main call and the 2nd call.
The default approach, for this Diagnose radiative forcings option, involves the model diagnosing the radiative forcing based on the flux-difference being between the advancing call including the radiative effects of the forcing agent (as usual), and the 2nd diagnostic call to the radiation setting that species' concentration to zero (or not including its effects in the radiative transfer calculations).
These additional settings for the diagnostic call are set in three of the sub-panels of this Section 1 - 2: Radiation part of the Rose GUI. First it can be seen that there are "SW second call" and "LW second call" panels that become active. There is also a separate Diagnostic forcing panel for setting the precise arrangement for this second call to the radiation. In this separate panel it is possible to individually (or multiply) select the CLASSIC aerosol types to diagnose their radiative effects. For GLOMAP-mode it's different in that it only makes sense to diagnose the effects over all the types considered since the different types are often internally mixed within each size class. The user will likely only want to run these effects when the settings in the second radiation call match exactly with those in the first radiation call (so that the difference then indicates the forcing due to the forcing agent of interest rather than any difference in settings). If particular experiments are planned, as usual it is recommended to discuss with the relevant expert(s) within NCAS or the Met Office.
As explained above, the default UM setting for the double-call is to set the species mixing ratio to zero in the diagnostic call with the difference then including any fast feedbacks from the forcing agent in the advancing call. However, it is often very useful to be able to suppress the fast feedbacks from the forcing agent in question by reversing the operation of the double-call including the aerosol radiative effects only in the diagnostic call and setting the species mixing ratio to zero in the advancing call.
With this diagnostic double-call radiative forcing configuration, the difference in radiative fluxes between the two calls then provides the aerosol radiative perturbation with respect to an atmosphere containing no aerosols. For example, one can diagnose the present-day to pre-industrial aerosol radiative forcing by taking the difference between two parallel diagnostic-double-call simulations with aerosol and precursor emissions set to 1850 and 2000. As long as other forcing agents, such as greenhouse gases or land-use change, remain fixed at a reference time period, the meteorology should then be identical in the two runs, giving a radiative forcing signal "clean" from the different transport arising from the dynamical response to the aerosol radiative effects.
Often nudging to meteorological re-analysis winds and temperatures is applied in tandem with the double-call configuration in which case the composition-climate model is being run in a similar way to an offline chemistry-transport model. This approach has been used extensively in aerosol forcing intercomparisons (e.g. the AeroCom direct forcing experiments, Myhre et al., 2013, ACP) with the radiative forcings diagnosed from each model with fast feedbacks disabled.
Unfortunately, this diagnostic double-call configuration is not one of the supported configurations of the v10.4 release job. As a consequence, to run UM-UKCA with this configuration, you will need to add an extra FCM branch to the job and also modify the metadata in the the Rose GUI to enable the option to run configured in this way.
First add the branch. In the fcm_make_um app go to env and then Sources. There you should add an extra entry to add-in the code-changes from revision 32452 of the following FCM branch:
branches/dev/grahammann/vn10.4_updates_for_dblcalaerforc@32452
The code added in that branch applies a new switch C2C_AER_DIAGCAL which controls whether the setting of the forcing agent to zero is applied on the advancing call (C2C_AER_DIAGCAL is .TRUE.) or to retain the standard operation of the double-call where the aerosol is included in the advancing call but set to zero in the diagnostic call (C2C_AER_DIAGCAL is .FALSE).
We need to use new metadata to enable this switch -- and also to add in a new switch C2C_UKCA_I which controls whether the GLOMAP indirect effects are included or not.
Just like in the last task, this requires to point the Rose GUI to a new rose-meta.conf file within a separate directory outside the FCM branch.
In the Rose GUI click on the um word and you'll see that there is already a separate metadata file specified in that job which is set to point to:
/home/grenville/meta/ga7_vn10.4
I have prepared a modified version of this that enables the extra C2C_AER_DIAGCAL and C2C_UKCA_I switches to be selected in the Rose GUI. So change the file path to instead point to this one:
/home/gmann/meta/ga7_vn10.4_dblcalaerforc
After you have done this, there will be 2 additional buttons available in the Diagnostic forcing panel to allow the user to specify the C2C_AER_DIAGCAL and C2C_UKCA_I switches to be true or false.
We want to use this diagnostic double call configuration and we want to have the forcing be including both the direct and indirect effects from the GLOMAP aerosol -- so set C2C_UKCA_D and C2C_UKCA_I to be .TRUE. You should also set the C2C_DUST_D switch to be true so that the model is then calculating the overall aerosol radiative effects (including both aerosol direct and indirect effects with the aerosol consisting of the total of the 6 CLASSIC dust bins plus the GLOMAP aerosol modes). All the other C2C switches in that panel should be set to .FALSE.
You will also need to update your job to make sure all the settings for the diagnostic call so that they match those set for the prognostic call (in terms of the methods for cloud water content variability, overlapping clouds, and which other forcing agents are interacting (or not).
To check this you should compare the "SW main call" and "SW second call" windows and the "LW main call" and the "LW second call" panels.
Firstly you need to add the spectral file name in both second call to match that used in the main call -- see the spectral files used are sp_sw_ga7 for the shortwave and sp_lw_ga7 for the longwave.
Next, select the Shortwave panel and "Number of times per day for full SW radiation calls" is set to 24, which means the radiation scheme is called every hour. The contrasts with the actual model timestep which is set at 20 minutes (see Top Level Model Control --> Model Domain and Timestep). In our double-call case, at every radiation timestep, we want there to be two calls. In other words the frequency for the second SW call needs to be set to match that in the full radiation call. It might seem the user would always want the two to match, but it turns out that in the so-called single-call timestepping configuration, this 2nd call is used to enable to call the radiation scheme more frequently but with a reduced operation of the radiation interactions (for details see Manners et al., QJRMS 2009). That's why the first call is referred to as a full call (because in the single-call timestepping configuration the second call is a reduced one. But in our double-call case, the second call is also full and we are diagnosing (on each radiation timestep) the difference to isolate the effects from the forcing agent of interest.
For the Longwave panel, also set the frequency to 24 per day for both.
All that remains then is to add in the extra STASH requests for the double-call forcing diagnostics which give the "increment" (flux-difference) between the two calls and to note a change to the way the AOD diagnostics operate.
The approach taken to index the STASH numbers for the radiative forcing items (the flux-difference between the two radiation calls) is to apply an offset of +200 to the item number to the corresponding item for the conventional radiative fluxes.
In task 12.1 we added STASH requests for the all-sky TOA outgoing SW and LW radiative fluxes which are referenced in STASH as section 1 item 208 and section 2 item 205.
To request the all-sky TOA outgoing SW and LW radiative forcings (between the two radiation calls) the corresponding item numbers are section 1 item 408 and section 2 item 405. Unfortunately however, at UM v10.4, section 2 item 405 is not available from the Rose GUI. In this task we will therefore request instead the clear-sky TOA outgoing SW and LW radiative forcing diagnostics (section 1 item 409 and section 2 item 406). Note also that one needs to request both the radiation flux and radiation forcing diagnostic in these runs so you should add 4 daily-mean STASH requests for section 1 items 209 and 409 and section 2 items 206 and 406.
Go to the STASH Specification of Diagnostic requirements window and add the daily-mean (TDAYM) requests for both of these flux-forcing pairs of diagnostics. As in Task 10.1, the domain profile should be set as DIAG and the usage profile as UPK. Note however that the TDAYM time profile is set to sample from the diagnostics on every timestep, whereas in fact these forcing diagnostics are only defined on radiation timesteps. As a consequence, we need to make up a new time profile TDMN_RAD which samples every 3 timesteps (on radiation timesteps)) instead of on every timestep. To do this requires to go to STASH requests and Profiles and select Time Profiles. Again, this is quite an intimidating panel of data, but actually it gives the UM a great deal of flexibility to sample diagnostics during runtime in a myriad of different ways. Here we want to take a copy of the existing TDAYM time profile (which calculated daily-means based on the average of values on every timestep) and instead change it to only sample the data every hour (ignoring the values on the other timesteps). You see there is a column in the Table entitled tim_name -- this should really be the first column of data because it tells you what the name of the profile is. You see there are profiles such as TDAYMAX -- that takes the maximum value over the day (useful for example in air quality applications for diagnosing exceedance days) and other useful ways of storing some statistics on the data as it is running. This flexibility does potentially make it seem difficult to understand. You can get an idea of what each time profile does by noting its index in the far left column. This number will be in the list underneath. For example the TDAYMAX time profile has the code 3ffa377f. When you then click on that code underneath a panel appears explaining exactly what it does. In our case, we want to take a copy of the TDAYM profile. This is done by right-clicking on the word TDAYM in the tim_name column and then selecting Clone this section. It's confusing because the word section is not really correct here -- it should say profile not section. Still, you know what it means. When you do that it generates a copy of the TDAYM and you should change that name to TDMN_RAD and then change the specified frequency of isam ("Specify the sampling period (frequency)") to be 3 instead of 1. See it says underneath that the units for the sampling period are set as timesteps. So we're changing the sampling frequency from once every timestep to once every three timesteps. Once this is done, it's just a case of saving the profile and requesting these double-call radiation diags (1-209, 1-409, 2-206, 2-406) with TDMN_RAD, DIAG and UPK.
Since we have set C2C_AER_DIAGCAL to be .TRUE., when the model runs it will be settings the GLOMAP aerosol to zero in the advancing call. So the conventional AOD diagnostics introduced in tutorial 10 (section 2 items 300 to 305) will now contain zero values when the model is run.
The UM therefore has a second set of AOD diagnostics giving the aerosol optical depth as calculated in the diagnostic call.
The approach for the double-call AOD diagnostics is the same as for the forcing diags, i.e. to apply an offset of +200 to the item number to find the corresponding AOD item in the diagnostic call.
The double-call GLOMAP AOD diagnostics are therefore found in section 2 items 500 to 505. I have added these extra STASH items to the modified STASHmaster_A in the ~gmann/meta/ga7_vn10.4_dblcalaerforc directory that the job is pointing to, so those double-call GLOMAP AODs will be available to select in the Rose GUI. Again, request these with the TDMN_RAD time profile, usage profile UPK and (this time) use domain profile DIAG_AOT (which specifies that 6 pseudo-levels are required for the 6 wavelengths the AODs are calculated for).
The simulation will then output daily-mean SW and LW clear-sky forcings and double-call AOD diagnostics to the .pk file for your v10.4 UM-UKCA job.
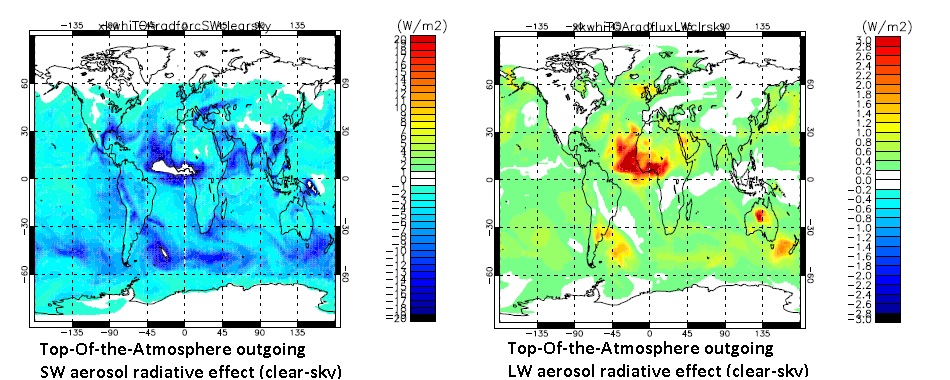
The Figure below shows daily-mean TOA SW-clearsky and LW-clearsky radiative effect fields for the double-call-modified version of the UKCA tutorial (gmann job u-ai978).
Worked Solution
A worked solution to Task 12.2 can be found in job u-ai978.
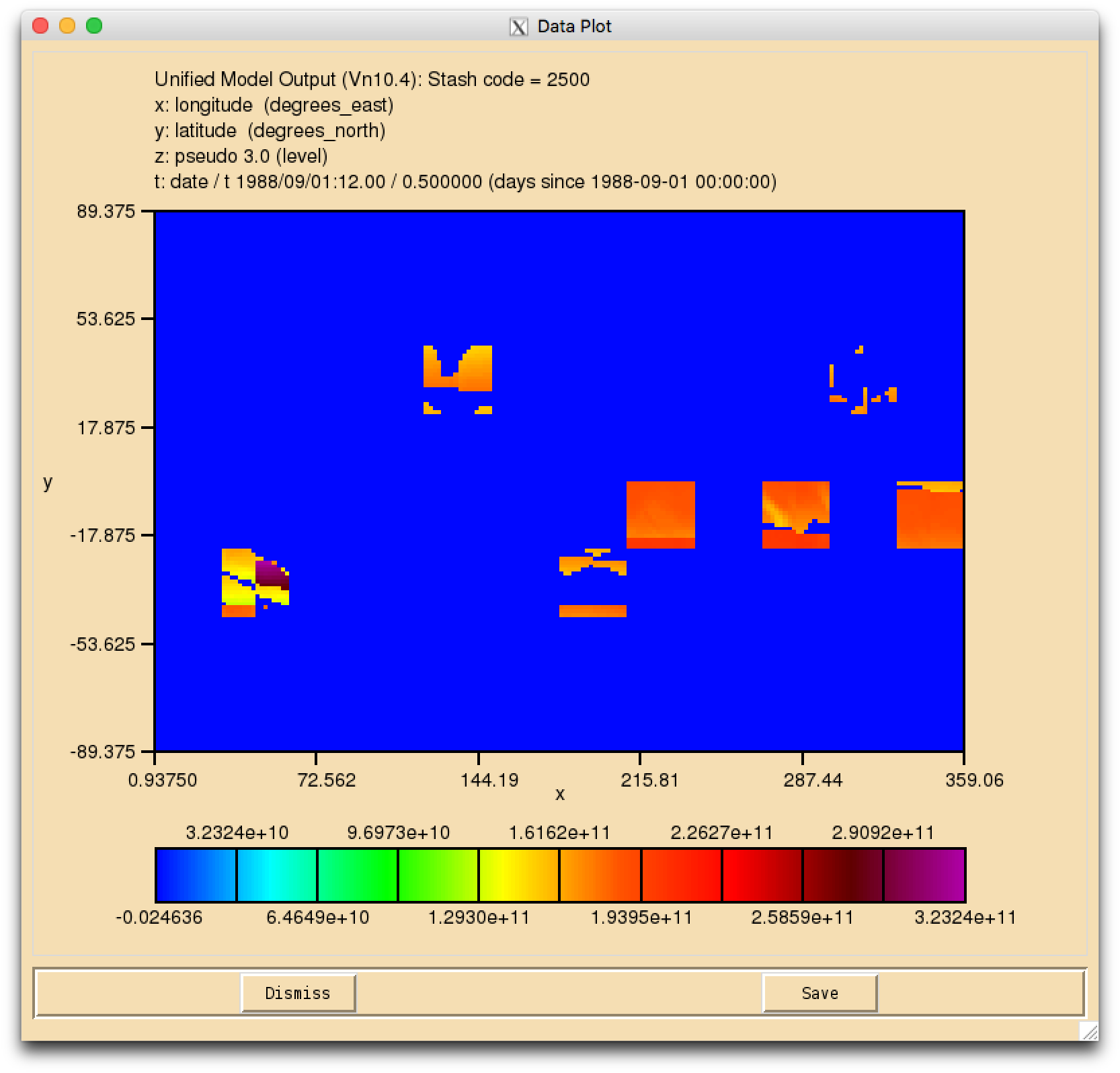
It should be noted that due to problems with setting up this configuration, the s02i500 to s02i505 fields are corrupted. To prevent the run from failing you need to change the packing of the .pk file to 'Unpacked (0)'. This can be done in um → namelist → Model Input and Output → Model Output Streams → pp10. With this done the file is created with the fields in place, but it is clear that the diagnostics are corrupted (see Figure 1).
It should be noted that these values may be different in your job. It is currently unclear why this issue exists, but we hope to fix it soon.
Sample output from a copy of this job can be found on ARCHER in the directory
/work/n02/n02/ukca/Tutorial/vn10.4/sample_output/Task12.2
Your .pk file should contain the following fields:
0 : 192 144 1 1 Upward solar 1 : 192 144 1 1 Clear-sky flux (type II) solar up 2 : 192 144 1 1 Clear-sky flux (type II) solar up 3 : 192 144 1 1 IR up 4 : 192 144 1 1 Clear-sky (type II) IR up 5 : 192 144 6 1 Unspecified 6 : 192 144 6 1 Unspecified 7 : 192 144 6 1 Unspecified 8 : 192 144 6 1 Unspecified 9 : 192 144 6 1 Unspecified 10 : 192 144 6 1 Unspecified 11 : 192 144 6 1 Unspecified 12 : 192 144 6 1 Unspecified 13 : 192 144 6 1 Unspecified 14 : 192 144 6 1 Unspecified 15 : 192 144 6 1 Unspecified 16 : 192 144 6 1 Unspecified 17 : 192 144 1 1 Clear-sky (type II) IR up 18 : 192 144 6 1 Unspecified 19 : 192 144 6 1 Unspecified 20 : 192 144 6 1 Unspecified 21 : 192 144 6 1 Unspecified 22 : 192 144 6 1 Unspecified 23 : 192 144 6 1 Unspecified 24 : 192 144 85 1 AITKEN MODE (SOLUBLE) OC MMR 25 : 192 144 85 1 ACCUMULATION MODE (SOL) OC MMR 26 : 192 144 85 1 COARSE MODE (SOLUBLE) OC MMR 27 : 192 144 85 1 AITKEN MODE (INSOLUBLE) OC MMR 28 : 192 144 85 1 NUCLEATION MODE (SOLUBLE) OC MMR
The Rose specific changes required for this task are (excluding the STASHmaster_A changes):
Index: app/um/rose-app.conf
===================================================================
--- app/um/rose-app.conf (revision 28651)
+++ app/um/rose-app.conf (revision 28652)
@@ -1,4 +1,4 @@
-meta=/home/grenville/meta/ga7_vn10.4
+meta=/home/gmann/meta/ga7_vn10.4_dblcalaerforc
[command]
default=um-atmos
@@ -23,6 +23,7 @@
RECON_KEEP_MPP_STDOUT=true
RECON_STDOUT_FILE=pe_output/${RUNID}.fort6.pe
SPECTRAL_FILE_DIR=$UMDIR/vn$VN/ctldata/spectral/ga7
+STASHMSTR=.
UM_THREAD_LEVEL=MULTIPLE
VN=10.4
@@ -2832,7 +2833,7 @@
!!filename='$ROSE_DATAC/$RUNID.pp10'
filename_base='$DATAM/${RUNID}a.pk%C'
l_reinit=.true.
-packing=5
+packing=0
reinit_end=-1
reinit_start=0
reinit_step=90
@@ -3004,140 +3005,142 @@
[namelist:r2lwclnl]
i_cnv_ice_lw=8
-!!i_cnv_ice_lw2=8
+i_cnv_ice_lw2=8
i_cnv_water_lw=5
-!!i_cnv_water_lw2=5
+i_cnv_water_lw2=5
i_gas_overlap_lw=4
-!!i_gas_overlap_lw2=6
+i_gas_overlap_lw2=4
i_scatter_method_lw=5
-!!i_scatter_method_lw2=1
+i_scatter_method_lw2=5
i_st_ice_lw=11
-!!i_st_ice_lw2=8
+i_st_ice_lw2=11
i_st_water_lw=5
-!!i_st_water_lw2=5
+i_st_water_lw2=5
l_cfc113_lw=.false.
-!!l_cfc113_lw2=.false.
+l_cfc113_lw2=.false.
l_cfc114_lw=.false.
-!!l_cfc114_lw2=.false.
+l_cfc114_lw2=.false.
l_cfc11_lw=.false.
-!!l_cfc11_lw2=.false.
+l_cfc11_lw2=.false.
l_cfc12_lw=.true.
-!!l_cfc12_lw2=.false.
+l_cfc12_lw2=.true.
l_ch4_lw=.true.
-!!l_ch4_lw2=.false.
+l_ch4_lw2=.true.
l_co2_lw=.true.
-!!l_co2_lw2=.true.
+l_co2_lw2=.true.
l_co_lw=.false.
-!!l_co_lw2=.false.
+l_co_lw2=.false.
l_cs_lw=.false.
-!!l_cs_lw2=.false.
+l_cs_lw2=.false.
l_extra_top_lw=.false.
-!!l_extra_top_lw2=.false.
+l_extra_top_lw2=.false.
l_h2_lw=.false.
-!!l_h2_lw2=.false.
+l_h2_lw2=.false.
l_hcfc22_lw=.false.
-!!l_hcfc22_lw2=.false.
+l_hcfc22_lw2=.false.
l_he_lw=.false.
-!!l_he_lw2=.false.
+l_he_lw2=.false.
l_hfc125_lw=.false.
-!!l_hfc125_lw2=.false.
+l_hfc125_lw2=.false.
l_hfc134a_lw=.true.
-!!l_hfc134a_lw2=.false.
+l_hfc134a_lw2=.true.
l_k_lw=.false.
-!!l_k_lw2=.false.
+l_k_lw2=.false.
l_li_lw=.false.
-!!l_li_lw2=.false.
+l_li_lw2=.false.
l_microphysics_lw=.true.
-!!l_microphysics_lw2=.false.
+l_microphysics_lw2=.true.
l_n2o_lw=.true.
-!!l_n2o_lw2=.false.
+l_n2o_lw2=.true.
l_na_lw=.false.
-!!l_na_lw2=.false.
+l_na_lw2=.false.
l_nh3_lw=.false.
-!!l_nh3_lw2=.false.
+l_nh3_lw2=.false.
l_rb_lw=.false.
-!!l_rb_lw2=.false.
+l_rb_lw2=.false.
l_solar_tail_flux=.false.
-!!l_solar_tail_flux_2=.false.
+l_solar_tail_flux_2=.false.
l_tio_lw=.false.
-!!l_tio_lw2=.false.
+l_tio_lw2=.false.
l_vo_lw=.false.
-!!l_vo_lw2=.false.
+l_vo_lw2=.false.
spectral_file_lw='sp_lw_ga7'
-!!spectral_file_lw2=
+spectral_file_lw2='sp_lw_ga7'
[namelist:r2swclnl]
i_cnv_ice_sw=8
-!!i_cnv_ice_sw2=8
+i_cnv_ice_sw2=8
i_cnv_water_sw=5
-!!i_cnv_water_sw2=5
+i_cnv_water_sw2=5
i_gas_overlap_sw=4
-!!i_gas_overlap_sw2=5
+i_gas_overlap_sw2=4
i_st_ice_sw=11
-!!i_st_ice_sw2=8
+i_st_ice_sw2=11
i_st_water_sw=5
-!!i_st_water_sw2=5
+i_st_water_sw2=5
l_ch4_sw=.true.
-!!l_ch4_sw2=.false.
+l_ch4_sw2=.true.
l_co2_sw=.true.
-!!l_co2_sw2=.true.
+l_co2_sw2=.true.
l_co_sw=.false.
-!!l_co_sw2=.false.
+l_co_sw2=.false.
l_cs_sw=.false.
-!!l_cs_sw2=.false.
+l_cs_sw2=.false.
l_extra_top_sw=.false.
-!!l_extra_top_sw2=.false.
+l_extra_top_sw2=.false.
l_h2_sw=.false.
-!!l_h2_sw2=.false.
+l_h2_sw2=.false.
l_he_sw=.false.
-!!l_he_sw2=.false.
+l_he_sw2=.false.
l_k_sw=.false.
-!!l_k_sw2=.false.
+l_k_sw2=.false.
l_li_sw=.false.
-!!l_li_sw2=.false.
+l_li_sw2=.false.
l_n2o_sw=.true.
-!!l_n2o_sw2=.false.
+l_n2o_sw2=.true.
l_na_sw=.false.
-!!l_na_sw2=.false.
+l_na_sw2=.false.
l_nh3_sw=.false.
-!!l_nh3_sw2=.false.
+l_nh3_sw2=.false.
l_o2_sw=.true.
-!!l_o2_sw2=.false.
+l_o2_sw2=.true.
l_rb_sw=.false.
-!!l_rb_sw2=.false.
+l_rb_sw2=.false.
l_solvar_sw=.false.
-!!l_solvar_sw2=.false.
+l_solvar_sw2=.false.
l_tio_sw=.false.
-!!l_tio_sw2=.false.
+l_tio_sw2=.false.
l_vo_sw=.false.
-!!l_vo_sw2=.false.
+l_vo_sw2=.false.
spectral_file_sw='sp_sw_ga7'
-!!spectral_file_sw2=
+spectral_file_sw2='sp_sw_ga7'
[namelist:radfcdia]
-!!c2c_aerosol=.false.
-!!c2c_all=.false.
-!!c2c_biog_d=.false.
-!!c2c_bmb_d=.false.
-!!c2c_c113=.false.
-!!c2c_cfc11=.false.
-!!c2c_cfc12=.false.
-!!c2c_ch4=.false.
-!!c2c_co2=.false.
-!!c2c_dust_d=.false.
-!!c2c_hcfc22=.false.
-!!c2c_hfc125=.false.
-!!c2c_hfc134=.false.
-!!c2c_land_s=.false.
-!!c2c_n2o=.false.
-!!c2c_nitr_d=.false.
-!!c2c_o3=.false.
-!!c2c_ocff_d=.false.
-!!c2c_seas_d=.false.
-!!c2c_soot_d=.false.
-!!c2c_sulpc_d=.false.
-!!c2c_ukca_d=.false.
-!!c2c_wmg=.false.
+c2c_aer_diagcal=.true.
+c2c_aerosol=.false.
+c2c_all=.false.
+c2c_biog_d=.false.
+c2c_bmb_d=.false.
+c2c_c113=.false.
+c2c_cfc11=.false.
+c2c_cfc12=.false.
+c2c_ch4=.false.
+c2c_co2=.false.
+c2c_dust_d=.true.
+c2c_hcfc22=.false.
+c2c_hfc125=.false.
+c2c_hfc134=.false.
+c2c_land_s=.false.
+c2c_n2o=.false.
+c2c_nitr_d=.false.
+c2c_o3=.false.
+c2c_ocff_d=.false.
+c2c_seas_d=.false.
+c2c_soot_d=.false.
+c2c_sulpc_d=.false.
+c2c_ukca_d=.true.
+c2c_ukca_i=.true.
+c2c_wmg=.false.
[namelist:recon]
ainitial='$AINITIAL_N96'
@@ -3743,19 +3746,19 @@
hfc125mmr=0.0
hfc134ammr=0
i_cloud_representation=2
-!!i_cloud_representation_2=2
+i_cloud_representation_2=2
i_fsd=2
-!!i_fsd_2=0
+i_fsd_2=0
i_inhom=2
-!!i_inhom_2=0
-!!i_lw_radstep_perday_diag=24
+i_inhom_2=0
+i_lw_radstep_perday_diag=24
i_lw_radstep_perday_prog=24
i_overlap=3
-!!i_overlap_2=0
+i_overlap_2=0
i_ozone_int=2
-i_rad_extra_call=0
+i_rad_extra_call=1
i_rad_topography=2
-!!i_sw_radstep_perday_diag=24
+i_sw_radstep_perday_diag=24
i_sw_radstep_perday_prog=24
inhom_cloud_lw=0
inhom_cloud_sw=0
@@ -4078,6 +4081,54 @@
ukcacrsw='$UMDIR/vn$VN/ctldata/UKCA/radaer/nml_cr_sw'
ukcaprec='$UMDIR/vn$VN/ctldata/spectral/ga7/RADAER_pcalc.ukca'
+[namelist:streq(1)]
+dom_name='DIAGAOT'
+isec=2
+item=500
+package=
+tim_name='TDMN_RAD'
+use_name='UPK'
+
+[namelist:streq(2)]
+dom_name='DIAGAOT'
+isec=2
+item=501
+package=
+tim_name='TDMN_RAD'
+use_name='UPK'
+
+[namelist:streq(3)]
+dom_name='DIAGAOT'
+isec=2
+item=502
+package=
+tim_name='TDMN_RAD'
+use_name='UPK'
+
+[namelist:streq(4)]
+dom_name='DIAGAOT'
+isec=2
+item=503
+package=
+tim_name='TDMN_RAD'
+use_name='UPK'
+
+[namelist:streq(5)]
+dom_name='DIAGAOT'
+isec=2
+item=504
+package=
+tim_name='TDMN_RAD'
+use_name='UPK'
+
+[namelist:streq(6)]
+dom_name='DIAGAOT'
+isec=2
+item=505
+package=
+tim_name='TDMN_RAD'
+use_name='UPK'
+
[namelist:streq(00687592)]
dom_name='DALLTH'
isec=38
@@ -4966,6 +5017,14 @@
tim_name='T6HDAYM'
use_name='UPB'
+[namelist:streq(1fdb379b)]
+dom_name='DIAG'
+isec=1
+item=409
+package=
+tim_name='TDMN_RAD'
+use_name='UPK'
+
[namelist:streq(200da117)]
dom_name='DIAG'
isec=1
@@ -6494,6 +6553,14 @@
tim_name='TDMPMN'
use_name='UPMEAN'
+[namelist:streq(5f71be2b)]
+dom_name='DIAG'
+isec=2
+item=406
+package=
+tim_name='TDMN_RAD'
+use_name='UPK'
+
[namelist:streq(5fb62131)]
dom_name='DP36CCMZ'
isec=6
@@ -7790,6 +7857,14 @@
tim_name='TDMPMN'
use_name='UPMEAN'
+[namelist:streq(989fcb6a)]
+dom_name='DIAG'
+isec=2
+item=206
+package=
+tim_name='TDMN_RAD'
+use_name='UPK'
+
[namelist:streq(98ace802)]
dom_name='DIAG'
isec=2
@@ -8750,6 +8825,14 @@
tim_name='TMPMN00'
use_name='UPMEAN'
+[namelist:streq(c305ce49)]
+dom_name='DIAG'
+isec=1
+item=209
+package=
+tim_name='TDMN_RAD'
+use_name='UPK'
+
[namelist:streq(c320ea17)]
dom_name='DIAG'
isec=2
@@ -10586,6 +10669,25 @@
!!unt2=2
unt3=2
+[namelist:time(a8072e80)]
+!!iedt=0
+iend=-1
+ifre=1
+intv=1
+ioff=0
+iopt=1
+isam=3
+!!isdt=0
+!!iser=0
+istr=1
+!!itimes=0
+ityp=3
+!!lts0=.false.
+tim_name='TDMN_RAD'
+unt1=3
+unt2=1
+unt3=3
+
[namelist:time(c34e59a7)]
!!iedt=0
iend=-1
Index: app/fcm_make_um/rose-app.conf
===================================================================
--- app/fcm_make_um/rose-app.conf (revision 28651)
+++ app/fcm_make_um/rose-app.conf (revision 28652)
@@ -45,4 +45,4 @@
stash_version=1A
timer_version=3A
um_rev=vn10.4
-um_sources=branches/dev/mohitdalvi/vn10.4_scale_lightning_nox@19623 branches/dev/stevenhardiman/vn10.4_ukca_tropopause_amendment@19627 branches/dev/alistairsellar/vn10.4_no_expvolc_so2@19808 branches/dev/marcuskoehler/vn10.4_ukca_fix_glomap_climatol_surfarea@24038
+um_sources=branches/dev/mohitdalvi/vn10.4_scale_lightning_nox@19623 branches/dev/stevenhardiman/vn10.4_ukca_tropopause_amendment@19627 branches/dev/alistairsellar/vn10.4_no_expvolc_so2@19808 branches/dev/marcuskoehler/vn10.4_ukca_fix_glomap_climatol_surfarea@24038 branches/dev/grahammann/vn10.4_updates_for_dblcalaerforc@32452
Index: rose-suite.conf
===================================================================
--- rose-suite.conf (revision 28651)
+++ rose-suite.conf (revision 28652)
@@ -2,7 +2,7 @@
!!ACCOUNT_USR='foundation'
ANCIL_OPT_KEYS=
ARCHER_GROUP='n02-training'
-ARCHER_QUEUE='short'
+ARCHER_QUEUE='standard'
!!ARCH_LOG=true
!!ARCH_WALL=true
ATM_PPN=24
These can also be found on PUMA here (including the STASHmaster_A changes):
/home/ukca/Tutorial/worked_solutions/Task12.2/task12.2.rose.diff
Written by Graham Mann 2016