UKCA Release Jobs: Difference between revisions
No edit summary |
|||
| (66 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
This page contains documentation on UKCA standard jobs that have |
This page contains documentation on UKCA standard jobs that have been released or will soon be released. |
||
'''You may also wish to look at the [[Bugfixes | bugfixes]] page.''' |
|||
==Getting Started== |
|||
'''You may also wish to look at the [[MONSooN IBM to Cray Transition]] page.''' |
|||
===PUMA=== |
|||
Please also see the [[Getting Started with UKCA]] page. |
|||
The UK Met Office's Unified Model (UM) is provided for the NERC community from NCAS Computational Modelling Services via the PUMA (Providing UM Access) resource. To use UKCA you must have an account on PUMA. To obtain one, please go to the [http://ncas-cms.nerc.ac.uk/index.php/puma NCAS-CMS PUMA page]. |
|||
===FCM=== |
|||
The UM uses code management software, built around [http://subversion.apache.org/ subversion], called [http://research.metoffice.gov.uk/research/nwp/external/fcm/doc/user_guide/ FCM] (Flexible Configuration Management). If you have not used the UM with FCM, we recommend first familiarising yourself with the system, by doing the [http://puma.nerc.ac.uk/trac/UM_TUTORIAL UM FCM Tutorial]. As well as being a good introduction to FCM and the UM User Interface (UMUI), it will also teach you how to set-up some configuration files that will be needed in the day-to-day running of the UM and UKCA. |
|||
==Standard Release Jobs== |
==Standard Release Jobs== |
||
UKCA can be configured through the UMUI, via it's own configuration panel. This mostly controls all the options required to run UKCA, although some extra changes have been made with hand-edits. To make it easy to get started with UKCA, several standard jobs have been created at various resolutions. To get started with UKCA, it should be a simple matter to take a copy of the UKCA job that you wish to run, changing a few user-specific options, and submitting the job. |
UKCA can be configured through the UMUI, via it's own configuration panel. This mostly controls all the options required to run UKCA, although some extra changes have been made with hand-edits. To make it easy to get started with UKCA, several standard jobs have been created at various resolutions. To get started with UKCA, it should be a simple matter to take a copy of the UKCA job that you wish to run, changing a few user-specific options, and submitting the job. |
||
Details of the jobs on ARCHER can be found [[ARCHER porting|here]]. |
|||
===Code Changes=== |
===Code Changes=== |
||
To run a UKCA standard job, please take a copy of one of the jobs listed below. If you need to make code changes, you must make a new FCM branch, and merge in the UKCA branch which is used in the job that you are taking, then use your branch instead. |
To run a UKCA standard job, please take a copy of one of the jobs listed below. If you need to make code changes, you '''must''' make a new FCM branch, and merge in the UKCA branch which is used in the job that you are taking, then use your branch instead. |
||
Please '''do not''' take a checkout of the UKCA branch used in the UMUI FCM panel (e.g. fcm:um_br/dev/luke/VN7.3_UKCA_CheM) and run from this as a working copy. If the ''fcm commit'' command is run from this directory it would commit any changes you have made to this branch. In this case, this would affect the branch for other UKCA users. |
Please '''do not''' take a checkout of the UKCA branch used in the UMUI FCM panel (e.g. fcm:um_br/dev/luke/VN7.3_UKCA_CheM) and run from this as a working copy. If the ''fcm commit'' command is run from this directory it would commit any changes you have made to this branch. In this case, this would affect the branch for other UKCA users. |
||
| Line 25: | Line 23: | ||
Available release jobs are: |
Available release jobs are: |
||
{| border="1" |
{| border="1" cellpadding="2" |
||
! UM Version ||UMUI Job-ID || Climate Model ||Horizontal Resolution ||Vertical Levels ||Model Top || Scheme Used |
! Release Job # || UM Version ||UMUI Job-ID || Climate Model ||Horizontal Resolution ||Vertical Levels ||Model Top || Scheme Used || FCM Branch || Revision Number || Documentation |
||
|- |
|- |
||
|7.3 ||HECToR: <br/> MONSooN: || QESM-A || N48 (2.5°×3.75°) || L60 || ~84km || Tropospheric Chemistry with Isoprene |
| RJ1.0 || 7.3 ||HECToR:'''xfvfa''','''xfvfc''' <br/> MONSooN: '''xfvfb''' <br/> ARCHER: '''xfvfd''' || QESM-A || N48 (2.5°×3.75°) || L60 || ~84km || Tropospheric Chemistry with Isoprene || fcm:um_br/dev/luke/VN7.3_UKCA_CheM || 13783 || [[#Tropospheric Chemistry with Isoprene - TropIsop|TropIsop]] |
||
|- |
|- |
||
|7.3 || || HadGEM3-A r2.0 || N96 (1.25°×1.875°) || L63 || ~40km || Tropospheric Chemistry with Isoprene |
| || 7.3 || || HadGEM3-A r2.0 || N96 (1.25°×1.875°) || L63 || ~40km || Tropospheric Chemistry with Isoprene || || || |
||
|- |
|- |
||
|7.3 ||HECToR: <br/> MONSooN: || QESM-A || N48 || L60 || ~84km || Stratospheric Chemistry |
| RJ2.0 || 7.3 ||HECToR:'''xfvga''','''xfvgc''' <br/> MONSooN:'''xfvgb''' <br/> ARCHER: '''xfvgd''' || QESM-A || N48 || L60 || ~84km || Stratospheric Chemistry || fcm:um_br/dev/luke/VN7.3_UKCA_CheM || 13783 || [[#Stratospheric Chemistry - StratChem|StratChem]] |
||
|- |
|- |
||
|7.3 || || HadGEM3-A r2.0 || N96 || L85 || 85km || Stratospheric Chemistry |
| || 7.3 || || HadGEM3-A r2.0 || N96 || L85 || 85km || Stratospheric Chemistry || || || |
||
|- |
|- |
||
|7.3 || || QESM-A || N48 || L60 || ~84km || Simple Tropospheric Chemistry with MODE Aerosol |
| || 7.3 || || QESM-A || N48 || L60 || ~84km || Simple Tropospheric Chemistry with MODE Aerosol || || || |
||
|- |
|- |
||
|7.3 || || |
| RJ3.0 || 7.3 ||HECToR: <br/> MONSooN:'''xfxlb''' || || N96 || L38 || ~40km || Simple Tropospheric Chemistry with MODE Aerosol ||fcm:um_br/dev/mdalvi/VN7.3_MODEBLN_Rad_Indir || 5953 || [[#Simple Tropospheric Chemistry with GLOMAP-mode aerosol - UKCA-MODE|UKCA-MODE]] |
||
|- |
|- |
||
|7.3 || || |
| RJ3.1 || 7.3 ||HECToR: <br/> MONSooN:'''xfxld''' || || N96 || L38 || ~40km || Simple Tropospheric Chemistry with MODE Aerosol (Nudged) || fcm:um_br/dev/mdalvi/VN7.3_MODEBLN_Rad_Indir fcm:um_br/dev/mdalvi/VN7.3_nudged_new || 5953 <br />5586 || [[#Simple Tropospheric Chemistry with GLOMAP-mode aerosol - UKCA-MODE|UKCA-MODE]] |
||
|- |
|- |
||
| RJ4.0 || 8.4 || ARCHER: '''xlavb'''<br/>MONSooN: '''xlava''' || GA4.0 || N96 || L85 || 85km || CheST chemistry with GLOMAP-mode aerosols || fcm:um_br/pkg/Config/vn8.4_UKCA || 18311 || [[Release Job RJ4.0|CheST+GLOMAP-mode]] |
|||
|- |
|||
| RJ5.0 || 11.0+ || See '''[[GA7.1 StratTrop suites]]''' table for nudged and free-running || GA7.1 || N96e || L85 || 85km || StratTrop (CheST) with GLOMAP-mode aerosol || Runs from the trunk with possible additional branches || || [[Release Job UM11.0]] |
|||
|} |
|} |
||
| Line 49: | Line 50: | ||
* '''User-id''', '''Mail-id''' (email address), and '''Tic-Code''': <br/> Model Selection -> User Information and Target Machine -> General Details <br/> ''The user-id is your username on the target machine (i.e. HECToR).<br/> The tic-code will be the HECToR charging code, e.g. n02-chem. This code can be found by logging into the [https://www.hector.ac.uk/safe/ HECToR SAFE] pages, and viewing your account information.'' |
* '''User-id''', '''Mail-id''' (email address), and '''Tic-Code''': <br/> Model Selection -> User Information and Target Machine -> General Details <br/> ''The user-id is your username on the target machine (i.e. HECToR).<br/> The tic-code will be the HECToR charging code, e.g. n02-chem. This code can be found by logging into the [https://www.hector.ac.uk/safe/ HECToR SAFE] pages, and viewing your account information.'' |
||
* '''Target machine root extract directory''' (UM_ROUTDIR): <br/> Model Selection -> FCM Configuration -> FCM Extract and Build directories and Output levels <br/> ''This needs to be the full path to |
* '''Target machine root extract directory''' (UM_ROUTDIR): <br/> Model Selection -> FCM Configuration -> FCM Extract and Build directories and Output levels <br/> ''This needs to be the '''full path''' to one of your directories on the target machine (HECToR/ARCHER /home directory or MONSooN /projects directory).'' |
||
See also: [[Using the UMUI]] for information on Optional UMUI changes such as changing the run length, archiving, emissions etc |
|||
===Optional UMUI Changes=== |
|||
==Tropospheric Chemistry with Isoprene - ''TropIsop''== |
|||
As well as the required changes above, you may also want to change the following, depending on your experiments: |
|||
:''See also [[Tropospheric_Chemistry_with_Isoprene|Tropospheric Chemistry with Isoprene description page]].'' |
|||
* Model '''start date''' and '''run length''': <br/> Model Selection -> Input/Output Control and Resources -> Start Date and Run Length Options <br/> '''''NOTE:''' For Stratospheric Chemistry, the CFC and other long-lived trace gas concentrations will be highly dependent on the date - you will need to think about your chemical tracer initial conditions if you change the start date of a Stratospheric Chemistry run.'' |
|||
* '''Reference date for meaning''': <br/> Model Selection -> Atmosphere -> Control -> Post processing, Dumping & Meaning -> Dumping and meaning <br/> ''If you have changed the start date (above), you may need to change the reference date. If you do not do this, climate meaning may not work. Climate means will only be produced for a date after the reference date.'' |
|||
* '''Archiving''': <br/> Model Selection -> Post Processing -> Main Switch + General Questions <br/> ''Currently archiving is switched '''ON''' for most jobs.<br/> On HECToR all model output will placed in a directory called '''archive''' which is a sub-directory of your run directory.<br/> ON MONSooN, all data is copied to '''/nerc/PROJECT/USERID/JOBID'''.<br/> Superseded model restart dumps will be deleted.'' |
|||
* '''UKCA Surface Emissions''' (as '''''user single-level ancillary file'''''): <br/> Model Selection -> Atmosphere -> Ancillary and input data files -> Climatologies and potential climatologies -> User single-level ancillary files and fields <br/> ''The emissions data-sets are taken from those used for AR5 integrations, and represent the year 2000. You may wish to change these emissions - to do this we recommend the use of [http://ncas-cms.nerc.ac.uk//index.php?option=com_wrapper&Itemid=274 Xancil], provided by NCAS-CMS.'' |
|||
* '''UKCA 3D Emissions''' (as '''''user multi-level ancillary file'''''): <br/> Model Selection -> Atmosphere -> Ancillary and input data files -> Climatologies and potential climatologies -> User multi-level ancillary files and fields <br/> ''The standard aircraft emissions data-set comes from the Aero2K project, and represents the year 2000. You may wish to change these emissions - to do this we recommend the use of [http://ncas-cms.nerc.ac.uk//index.php?option=com_wrapper&Itemid=274 Xancil], provided by NCAS-CMS.'' |
|||
* '''Tracer Initial Conditions''': <br/> Model Selection -> Atmosphere -> STASH -> Initialisation of User Prognostics <br/> ''The table in this UMUI panel gives a listing of all the initial conditions of the STASH items defined by the user. They are given a STASH code, which the UM uses to identify the field. The UKCA tracers have a 5 digit code beginning at '''34001''' and ending at '''34150''', although 34149 and 34150 need to be initialised to zero. Several (or all) tracers will be initialised to some initial conditions, contained in the file listed by each tracer.'' |
|||
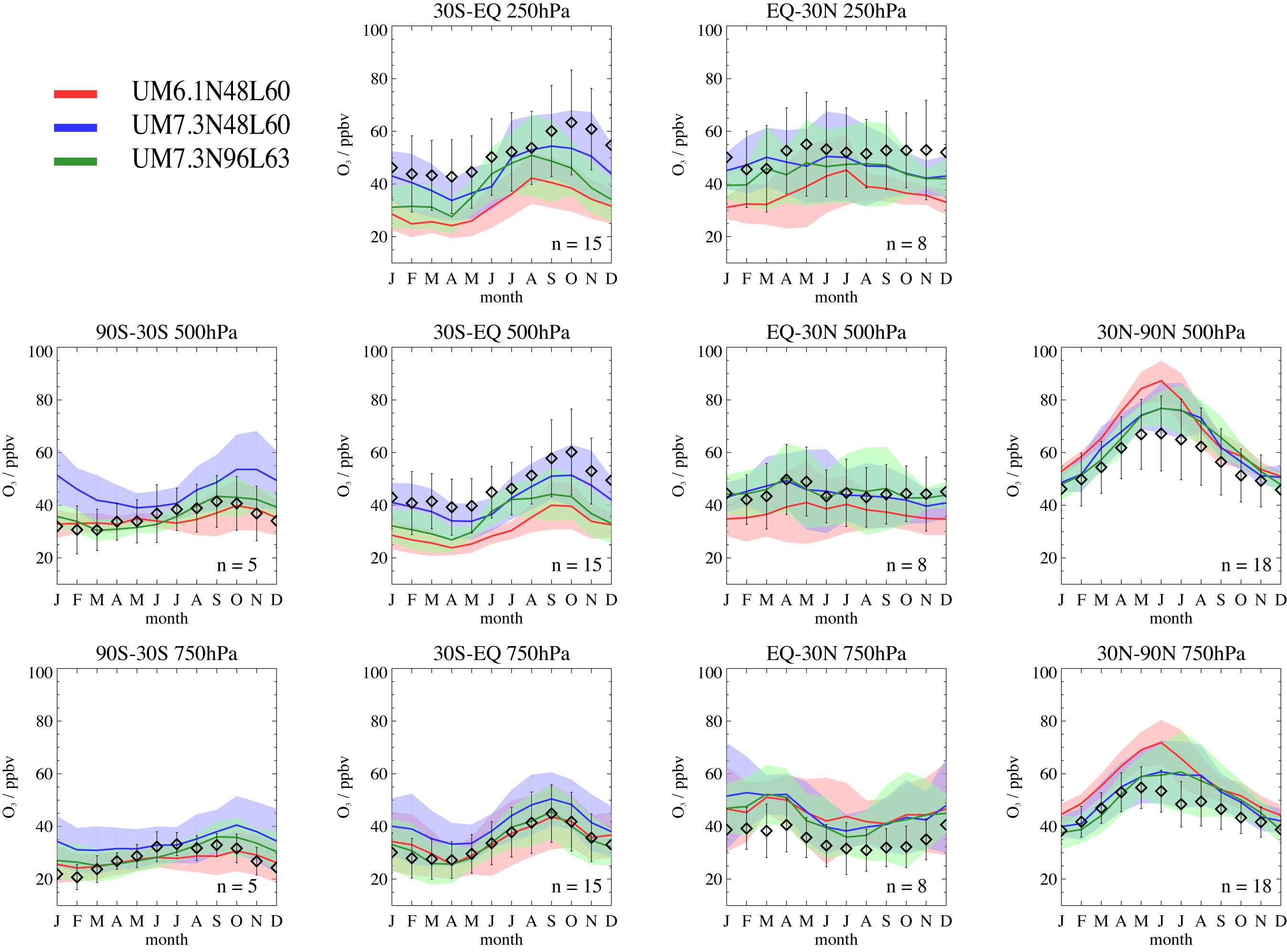
[[Image:TropIsop_comparison01.png|thumb|right|Comparison of Tropospheric Chemistry with Isoprene in a number of model resolutions and UM versions.]] This scheme using the FCM branch '''fcm:um_br/dev/luke/VN7.3_UKCA_CheM''' at '''revision 13783''' (''updated''). It has 56 species (49 of which are tracers), 115 bimolecular reactions, 35 photolysis reactions (using rates determined using a 2D look-up table) and 15 termolecular reactions. 23 species are wet-deposited and 32 are dry-deposited (using a 2D dry-deposition scheme). Tropospheric <math>O_{x}</math> and <math>CO</math> budgets are outputted as standard, as are other diagnostics of interest, such as the <math>CH_{4}</math> lifetime and Stratosphere-Troposphere Exchange of <math>O_{3}</math>. |
|||
====Continuation Run==== |
|||
The chemistry is integrated using the ASAD chemical package, using a sparse-matrix Newton-Raphson solver. While the UM dynamical timestep is every 20 model minutes, chemistry is called every model hour, although emissions are still performed every dynamical timestep. Top boundary conditions are used for <math>O_{3}</math> and <math>NO_{y}</math> and <math>CH_{4}</math> is treated as a constant. The tracers are initialised to spun-up fields from a previous ''TropIsop'' run, however, we recommend that a 15 month-spin-up should be performed. |
|||
After the job has run the first step, you will probably wish to turn on a continuation run. |
|||
The chemistry scheme used in these simulations is the same as in [http://www.atmos-chem-phys.net/10/7117/2010/acp-10-7117-2010.html Telford ''et al''], ''Atmos. Chem. Phys.'', '''10''', 7117-7125, doi:10.5194/acp-10-7117-2010 (2010). A model description paper is also in preparation (O'Connor ''et al'', 2011). |
|||
* '''Turn off compilation''': <br/> Model Selection -> Compilations and Modifications -> Compile options for the UM model <br/> Model Selection -> Compilations and Modifications -> Compile options for the UM reconfiguration <br/> ''Set to '''Run from existing executable, as named below''' in both panels'' |
|||
* Set to '''CRUN''': <br/> Model Selection -> Input/Output Control and Resources -> User hand edit files <br/> ''Find the hand-edit '''crun.ed''' and set this to '''Y''''' |
|||
The tropospheric ozone concentrations in UKCA-TropIsop are biased slightly high. This is improved with using an interactive photolysis scheme, which will be released for general use soon. |
|||
====Running a Perpetual Year==== |
|||
===Timings=== |
|||
For some experiments, you may wish to run a perpetual year (i.e. a simulation of multiple years in length, each one representing the same year). There are two ways to two this: |
|||
It should be noted that due to "jitter" on the HPC platform, all timings are approximate. As a simulation progresses the time per model month may increase as the fields spin-up from their initial state. Please also note that the MONSooN timings are for phase 1 - jobs will run more quickly on phase 2 and you should consider using more nodes than on phase 1. |
|||
# Manually re-run the same year |
|||
#* In this case you would run up until you have the 1st January start dump, then manually restart the simulation from this dump, run for 1 year and then send a new job off, running from the new 1st January start dump. You would need to ensure that the date used for each of these steps is for the year that you are interested in. To do this you will need to change how reconfiguration is performed. |
|||
#* '''Reconfiguration''': <br/> Model Selection -> Atmosphere -> Ancillary and input data files -> Start Dump <br/> ''You will need to alter the directory path and name of file to point to the start-dump you are interested in. You must then turn '''off''' the radio button '''Using the reconfiguation''' and make sure that the '''start-date''' (listed at the top of this panel) matches that in the dump file.'' |
|||
# Create a multi-year simulation where all the input files are for a single year |
|||
#* This method is particularly useful when you want to run a perpetual year for more than a few years, as might be the case for Stratospheric Chemistry when 15-20 year spin-ups are needed. There are a few changes that need to be made to several ancillary files and the UMUI: |
|||
## Change Ancillary files: |
|||
##* The following acillary files will need to be changed, since these are not climatologies by default, but time-series. |
|||
##** The '''single-level''' and '''multi-level''' (see above) user ancillary files may need to be changed to the year you are interested in. |
|||
##** '''Sea-Surface Temperature''': <br/> Model Selection -> Atmosphere -> Ancillary and input data files -> Climatologies & potential climatologies -> Sea surface temperatures |
|||
##** '''Sea-Ice''': <br/> Model Selection -> Atmosphere -> Ancillary and input data files -> Climatologies & potential climatologies -> Sea ice fields |
|||
##** '''2D Sulphur Emissions''': <br/> Model Selection -> Atmosphere -> Ancillary and input data files -> Climatologies & potential climatologies -> Sulphur cycle emissions (2D) <br/> ''NOTE: you should not need to change the DMS emissions'' |
|||
##** '''Soot Emissions''': <br/> Model Selection -> Atmosphere -> Ancillary and input data files -> Climatologies & potential climatologies -> Soot Emissions |
|||
##** '''Biomass Emissions''': <br/> Model Selection -> Atmosphere -> Ancillary and input data files -> Climatologies & potential climatologies -> Biomass Emissions |
|||
##* To create perpetual-year ancillary files: |
|||
##*# For each of the the ancillary files specified in these UMUI panels above, open in [http://badc.nerc.ac.uk/help/software/xconv/ Xconv] and select only the year that you are interested in and export this as a NetCDF file. |
|||
##*# Use [http://ncas-cms.nerc.ac.uk//index.php?option=com_wrapper&Itemid=274 Xancil] to create replacement ancillary files for each of the above files. You will need to make sure that these are made as ''periodic in time''. |
|||
##*# In the UMUI, replace the time-series ancillary files with your climatologies. |
|||
## UKCA changes: |
|||
##* The changes to UKCA are controlled by two hand-edits: |
|||
##*# '''CheM_aerosol_background_on.ed/CheM_aerosol_background_off.ed''' |
|||
##*#* This hand-edit either specifies using a background aerosol climatology for all years ('''_on'''), or using a time-series of atmospheric aerosols ('''_off'''). The default used is ''CheM_aerosol_background_off.ed''. |
|||
##*# '''CheM_trgas_presentday_full.ed''' (a pre-industrial version is also supplied, ''CheM_trgas_preI_full.ed'') |
|||
##*#* This hand-edit gives values of certain trace-gases to UKCA. Some are taken from the UMUI - these are: |
|||
##*#** <math>CH_{4}</math> |
|||
##*#** <math>N_{2}O</math> |
|||
##*#** <math>CFC11</math> (<math>CFCl_{3}</math>) |
|||
##*#** <math>CFC12</math> (<math>CF_{2}Cl_{2}</math>) |
|||
##*#** <math>CFC113</math> (<math>CF_{2}ClCFCl_{2}</math>) |
|||
##*#** <math>HCFC22</math> (<math>CHF_{2}Cl</math>) |
|||
##*#* Some gases are used by UKCA to specify constant values for the chemical solver. These are: |
|||
##*#** <math>CH_{4}</math> (''TropIsop'' chemistry only) |
|||
##*#** <math>CO_{2}</math> |
|||
##*#** <math>H_{2}</math> |
|||
##*#** <math>N_{2}</math> |
|||
##*#** <math>O_{2}</math> |
|||
##*#* Some trace-gases are used in Stratospheric Chemistry, to specify the surface concentration of certain UKCA species (<math>N_{2}O</math>, <math>CF_{2}Cl_{2}</math>, <math>CFCl_{3}</math>, <math>CH_{3}Br</math>, <math>H_{2}</math>, and <math>CH_{4}</math>). Some species are lumped together, so the surface concentrations of the following species can be set by the hand-edit: |
|||
##*#** <math>CH_{3}Br</math> (as ''MeBr'') |
|||
##*#** <math>CH_{3}Cl</math> (as ''MeCl'') |
|||
##*#** <math>CH_{2}Br_{2}</math> |
|||
##*#** <math>H_{2}</math> |
|||
##*#** <math>N_{2}</math> |
|||
##*#** <math>CFC114</math> |
|||
##*#** <math>CFC115</math> |
|||
##*#** <math>CCl_{4}</math> |
|||
##*#** <math>CH_{3}CCl_{3}</math> (as ''MeCCl3'') |
|||
##*#** <math>HCFC141b</math> |
|||
##*#** <math>HCFC142b</math> |
|||
##*#** <math>H1211</math> |
|||
##*#** <math>H1202</math> |
|||
##*#** <math>H1301</math> |
|||
##*#** <math>H2402</math> |
|||
##*#* To allow the specification of these gases in this way to be uses by UKCA, you will also need to turn on this option in the UMUI: '''UMUI values for CFCs etc''' <br/> Model Selection -> Atmosphere -> Model Configuration -> UKCA Chemistry and Aerosols -> STRAT |
|||
{| border="1" cellpadding="2" |
|||
====Adding New UKCA Tracers==== |
|||
! Machine ||No. of Cores ||No. of Nodes ||Resolution ||Time for 1 Model Month |
|||
|- |
|||
|HECToR Phase2b|| 96 || 4 || N48L60 || ~50 minutes |
|||
|- |
|||
|MONSooN Pahse 1|| 64 || 2 || N48L60 || ~59 minutes |
|||
|- |
|||
|ARCHER|| 72 || 3 || N48L60 || ~27 minutes |
|||
|- |
|||
|} |
|||
==Stratospheric Chemistry - ''StratChem''== |
|||
You may wish to add new UKCA tracers, possibly with associated reactions. This is possible for both the ''TropIsop'' and ''StratChem'' configurations. There are two steps that you need to do - first, set up the new tracers for the UM, and second set up these tracers for UKCA. Adding tracers will require both UMUI hand-edit, STASH, and source-code changes. |
|||
:''See also [[Standard_Stratospheric_Chemistry|Stratospheric Chemistry description page]].'' |
|||
# UMUI Changes |
|||
#* While these jobs only use a small sub-set of the total number of tracers allowed by the UM (150), due to the number of different schemes used, many of these other slots will have been reserved. While it is possible to use them, it is recommended that you choose an empty slot. To find which tracer slots are free, look in the file '''ukca_setd1defs.F90''' which is found in the ''src/atmosphere/UKCA'' directory. In this file, locate the array specification of '''nm_spec''' (''non-families''). This array contains all 150 currently considered tracers. Several of these slots are unused, and these are the ones labeled ''XXX''. Work out which number these are in the array, e.g. in the VN7.3_UKCA_CheM branch, numbers 40, 60, and 96-100 are currently free. |
|||
#* Take a copy of the tracer pre-STASHmasterfile (PSM) used by the UKCA job, and add a section for the tracer you are adding. You will need to change 3 numbers and the tracer name: |
|||
#*# The ''item number'' ('''Item'''): line 1, entry 3 (should be 1-150) |
|||
#*# The ''option code'' ('''Option Codes'''): line 3, entry 1 (last 3 digits, same number as ''Item'') |
|||
#*# The ''field code'' ('''PPFC''') : line 5, entry 2 (tracers begin numbering at 501 for tracer 1) |
|||
#*# The ''name'' of the tracer ('''Name'''): line 1, entry 4 (this should be representative of the species you are adding for clarity) |
|||
#** An example of a PSM entry is [[UKCA_Release_Jobs#Blank_pre-STASHmasterfile|below]], with '''XXX''' marking the locations to be changed |
|||
#** You will need to make sure that the lengths of the lines remains the same, otherwise the UMUI will not be able to read the PSM. |
|||
#** Replace the previous tracer PSM with your new one in the UMUI STASHmaster Panel: <br/> Model Selection -> Atmosphere -> STASH -> User-STASHmasterfiles. Diags, Progs & Ancils. |
|||
#** Initialise this tracer to a value in the UMUI Initialisation Panel:<br/> Model Selection -> Atmosphere -> STASH -> Initialisation of User Prognostics <br/> ''Scroll down the list until you come to your new tracer. The '''Option''' column will be blank - either'' |
|||
#**# ''initialise to zero (option 3)'' |
|||
#**# ''initialise to a constant value (option 6, value specified in column 4)'' |
|||
#**# ''initialise to a 3D field (option 7, file containing field specified in column 5. You may wish to use [http://ncas-cms.nerc.ac.uk//index.php?option=com_wrapper&Itemid=274 Xancil] for this option)'' |
|||
#*** It may be best to set the tracer to zero or a constant value at this point. |
|||
#*** You may also wish to add the tracer through STASH to the climate meaning stream:<br/> Model Selection -> Atmosphere -> STASH -> STASH. Specification of Diagnostic requirements |
|||
#* With your new PSM, run the script '''make_tracer_list''', found in the ''/home/ukca/bin'' directory on PUMA on this file. The output should be similar to:<br/> <tt>TC_UKCA=1,1,1,1,1,1,1,1,0,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,<br/> 1,0,1,1,1,1,1,1,1,1,1,1,1,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,0,0,0,0,<br/> 0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,1,1,1,1,1,1,1,1,1,0,0,0,1,1,0,0,0,0,0,0,0,<br/> 0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,<br/> 0,0,0,0,0,0,0,0,1,</tt> |
|||
#** Take a copy of the hand-edit that your job uses to add the UKCA tracers. For ''TropIsop'' jobs this will be '''add_s34_CheT_tracers_L60/63.ed''' and for ''StratChem'' jobs this will be'''add_s34_CheS_tracers_L60/85.ed'''. |
|||
#**# Find the lines that have the '''TC_UKCA''' definition, and replace it with the line you have generated with your PSM. |
|||
#**# Find the lines that define the value of '''TR_UKCA''' and increment this by the number of tracers you are adding. |
|||
#** Replace the original hand-edit in the UMUI with the one you have just created: <br/> Model Selection -> Input/Output Control and Resources -> User hand edit files |
|||
#* Make sure '''reconfiguration''' is on: <br/> Model Selection -> Atmosphere -> Ancillary and input data files -> Start Dump <br/> ''The radio-button '''Using the reconfiguration''' should be '''on''''' |
|||
#* ''SAVE'', ''PROCESS'' and ''SUBMIT'' the job. This should run with the extra tracer added. Look in a dump file to be sure that the tracer is there, and that it's value is as was set in the initialisation panel. |
|||
# UKCA code changes |
|||
## Adding the tracer |
|||
##* Before making these changes, you should run through the [http://puma.nerc.ac.uk/trac/UM_TUTORIAL UM FCM Tutorial]. |
|||
##* Any code changes should be made to a branch that you own. Make a new branch and merge in the UKCA branch that you are using (e.g. ''fcm:br/dev/luke/VN7.3_UKCA_CheM'') |
|||
##* For these changes, you must be a copy of the UKCA branch. We suggest using a '''working copy''' rather than committing the changes: <br/> Model Selection -> FCM Configuration -> FCM Options for Atmosphere and Reconfiguration <br/> ''Set the UKCA branch to '''N''' and select the radio-button '''Include modifications from user working copy''' and put the '''full path''' (on PUMA) '''to the local copy of your branch''''' |
|||
##* Edit the '''nm_spec''' (non-Families) array so that the slot used for the PSM now contains your tracer |
|||
##* Take a copy of the '''jp_for_CheT/S.ed''' (<math>T=TropIsop</math>, <math>S=StratChem</math>) hand-edit: <br/> Model Selection -> Input/Output Control and Resources -> User hand edit files |
|||
##** Increase the values of '''JPCTR''' and '''JPSPEC''' by the number of tracers you are adding |
|||
##** Add the tracers: |
|||
##*** For '''StratChem''': Edit the file '''ukca_chem1_dat.F90''' to add your tracer(s) into the '''chch_defs_strat''' type array, making sure that you use the same name as in the ''nm_spec'' array. |
|||
##*** For '''TropIsop''': Edit the file '''ukca_chem1_tropisop.F90''' to add your tracer(s) into the '''chch_defs_tropisop''' type array, making sure that you use the same name as in the ''nm_spec'' array. |
|||
##* This should now run with the extra tracer considered as such by UKCA. However, there are no reactions/deposition being performed on it, so the tracer still should not change when viewed in a dump file |
|||
## Adding reactions, deposition etc |
|||
##* You will need to edit the '''ukca_chem1_dat.F90''' or '''ukca_chem1_tropisop.F90''' as appropriate to add reactions. These are defined |
|||
##** '''ratb_defs''': Bimolecular reactions ('''JPBK''') |
|||
##** '''ratt_defs''': Termolecular reactions ('''JPTK''') |
|||
##** '''rath_defs''': Heterogeneous reactions (for ''StratChem'' only) ('''JPHK''') |
|||
##** '''ratj_defs''': Photolysis reactions ('''JPPJ''') |
|||
##** '''depvel_defs''': Dry-deposition ('''JPDD''') |
|||
##*** For species which are dry-deposited, you will also need to edit the '''chch_defs''' type, and put a '''1''' in the first column of the block of three numbers |
|||
##*** The ordering of the ''depvel_defs'' type is dependent on the ordering of the tracers in ''chch_defs'', so you will need to make sure that this is correct |
|||
##** '''henry_defs''': Wet-deposition (Henry's Law) ('''JPDW''') |
|||
##*** For species which are wet-deposited, you will also need to edit the '''chch_defs''' type, and put a '''1''' in the second column of the block of three numbers |
|||
##*** The ordering of the ''henry_defs'' type is dependent on the ordering of the tracers in ''chch_defs'', so you will need to make sure that this is correct |
|||
##** '''''NOTE:''' Emissions are not considered here'' |
|||
##** These configurations of UKCA are built using the ASAD chemical integration package. For documentation on using ASAD, please see the [http://www.atm.ch.cam.ac.uk/acmsu/asad/ ASAD webpage]. |
|||
##* Edit the '''jp_for_CheT/S.ed''' to increment the '''JP''' counter(s) (listed above) associated with your changes, and also increase the size of '''JPNR''', the total number of reactions, accordingly (dry- and wet-deposition are not counted in JPNR). |
|||
##* ''SAVE'', ''PROCESS'' and ''SUBMIT'' the job. This should now be using the new tracer in UKCA. |
|||
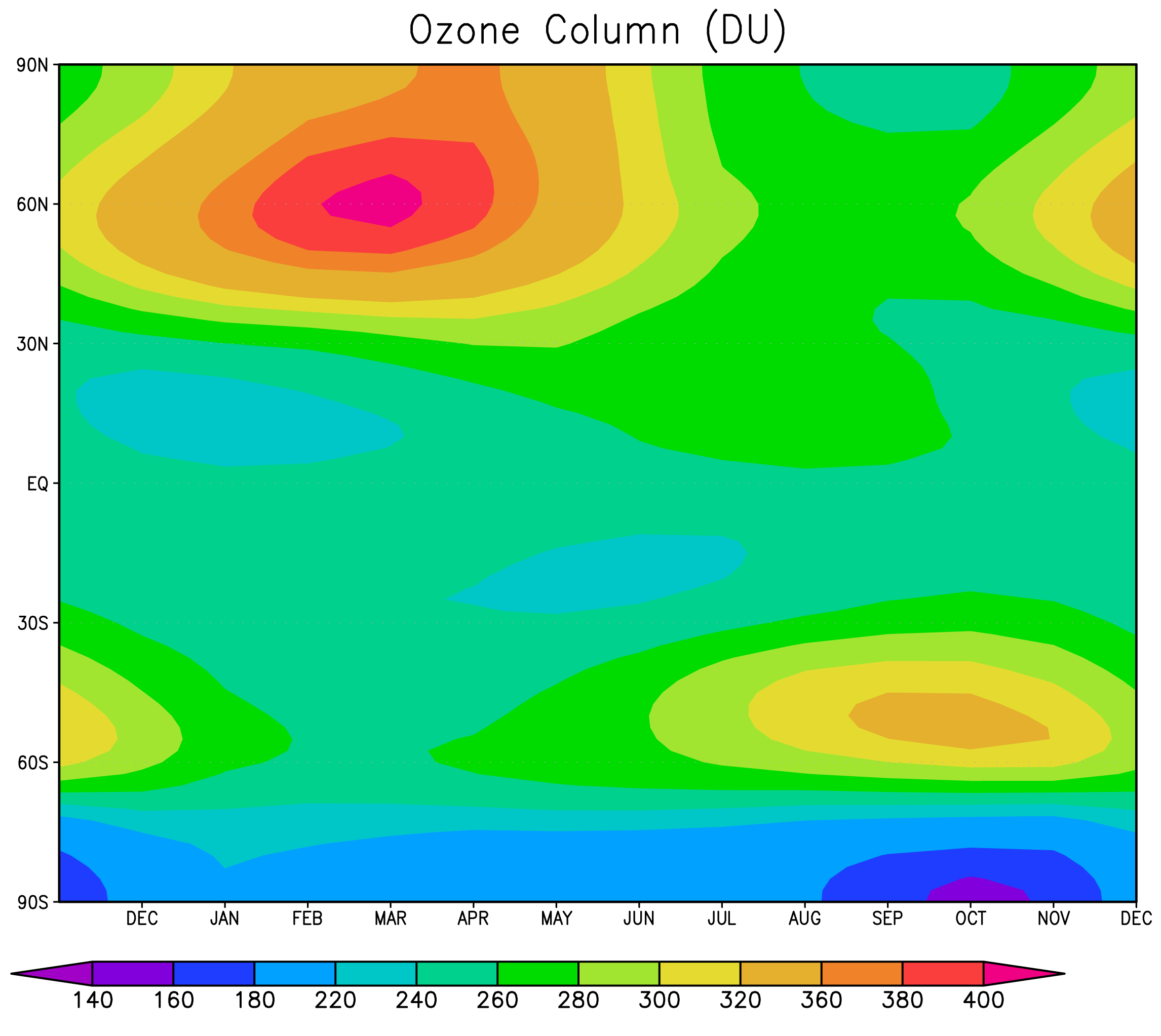
[[Image:O3_column.png|thumb|right|Ozone column in Dobson Units (DU) calculated from a UM7.3 N48L60 UKCA-StratChem run using interactive UKCA ozone in the UM radiation scheme. This is the mean of 21 model years representing 1990.]] This scheme using the FCM branch '''fcm:um_br/dev/luke/VN7.3_UKCA_CheM''' at '''revision 13783''' (''updated''). It has 41 species (36 of which are tracers), 113 bimolecular reactions, 34 photolysis reactions (using rates determined using a 3D look-up table), 17 termolecular reactions and 5 heterogeneous reactions. 15 species are wet-deposited and 15 are dry-deposited (using a 2D dry-deposition scheme). A stratospheric <math>O_{3}</math> is standard (based on the budget of Lee ''et al'', ''JGR'' '''107''' [http://www.agu.org/journals/ABS/2002/2001JD000538.shtml DOI:10.1029/2001JD000538] (2002)), as are other diagnostics of interest, such as the <math>CH_{4}</math> lifetime and Stratosphere-Troposphere Exchange of <math>O_{3}</math>. |
|||
=====Blank pre-STASHmasterfile===== |
|||
The chemistry is integrated using the ASAD chemical package, using a sparse-matrix Newton-Raphson solver. While the UM dynamical timestep is every 20 model minutes, chemistry is called every model hour, although emissions are still performed every dynamical timestep. By default, lower boundary conditions set for the tracer concentrations of <math>N_{2}O</math>, <math>CH_{4}</math>, <math>H_{2}</math>, <math>CF_{2}Cl_{2}</math>, <math>CFCl_{3}</math>, <math>CH_{3}Br</math>, and <math>CH_{2}Br_{2}</math> are specified for the years 1950-2100. These values are also used in the UM radiation scheme, along with <math>CO_{2}</math> concentrations. The tracers are initialised to spun-up fields from a previous ''StratChem'' run, however it can take time for a Stratospheric run to equilibrate and spin-up times as long as 20 model years may be required if significant changes are made. |
|||
The '''XXX''' labeled sections are ones which should be changed when [[UKCA_Release_Jobs#Adding_New_UKCA_Tracers|adding a new UKCA tracer]]. |
|||
The chemistry scheme used in these simulations is the similar to that described in [http://www.geosci-model-dev.net/2/43/2009/gmd-2-43-2009.html Morgenstern ''et al''], ''Geosci. Model Dev.'', '''2''', 43-57, (2009). |
|||
1| 1 | 34 | '''XXX''' | '''XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX''' | |
|||
2| 2 | 0 | 1 | 1 | 2 | 10 | 11 | 0 | 0 | 0 | 0 | |
|||
3| 000000000000000000000000000'''XXX''' | 00000000000000000001 | 1 | |
|||
4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | |
|||
5| 0 | '''XXX''' | 1 | 65 | 0 | 0 | 0 | 0 | 0 | |
|||
Please also see the [[Bugfixes#Total_N_tracer_initial_condition | bugfixes ]] page for further information on the initial conditions used in the release job. |
|||
====UMUI UKCA Panel==== |
|||
The settings in the UMUI UKCA panel have been chosen for that specific chemistry/aerosol scheme, and choosing other options may not work. If you require extra functionality, please contact [mailto:Luke.Abraham@atm.ch.cam.ac.uk Luke Abraham] to check on the current code status. |
|||
==Tropospheric Chemistry with Isoprene - ''TropIsop''== |
|||
[[Image:TropIsop_comparison01.png|thumb|right|Comparison of Tropospheric Chemistry with Isoprene in a number of model resolutions and UM versions.]] This scheme using the FCM branch '''fcm:um_br/dev/luke/VN7.3_UKCA_CheM''' at '''revision 5703'''. It has 56 species (49 of which are tracers), 115 bimolecular reactions, 35 photolysis reactions (using rates determined using a 2D look-up table) and 15 termolecular reactions. 23 species are wet-deposited and 32 are dry-deposited (using a 2D dry-deposition scheme). Tropospheric <math>O_{x}</math> and <math>CO</math> budgets are outputted as standard, as are other diagnostics of interest, such as the <math>CH_{4}</math> lifetime and Stratosphere-Troposphere Exchange of <math>O_{3}</math>. |
|||
The chemistry is integrated using the ASAD chemical package, using a sparse-matrix Newton-Raphson solver. While the UM dynamical timestep is every 20 model minutes, chemistry is called every model hour, although emissions are still performed every dynamical timestep. Top boundary conditions are used for <math>O_{3}</math> and <math>NO_{y}</math> and <math>CH_{4}</math> is treated as a constant. The tracers are initialised to spun-up fields from a previous ''TropIsop'' run, however, we recommend that a 15 month-spin-up should be performed. |
|||
The chemistry scheme used in these simulations is the same as in [http://www.atmos-chem-phys.net/10/7117/2010/acp-10-7117-2010.html Telford ''et al''], ''Atmos. Chem. Phys.'', '''10''', 7117-7125, doi:10.5194/acp-10-7117-2010 (2010). A model description paper is also in preparation (O'Connor ''et al'', 2011). |
|||
The tropospheric ozone concentrations in UKCA-TropIsop are biased slightly high. This is improved with using an interactive photolysis scheme, which will be released for general use soon. |
|||
===Timings=== |
===Timings=== |
||
It should be noted that due to "jitter" on the HPC platform, all timings are approximate. As a simulation progresses the time per model month may increase as the fields spin-up from their initial state. |
It should be noted that due to "jitter" on the HPC platform, all timings are approximate. As a simulation progresses the time per model month may increase as the fields spin-up from their initial state. Please also note that the MONSooN timings are for phase 1 - jobs will run more quickly on phase 2 and you should consider using more nodes than on phase 1. |
||
{| border="1" |
{| border="1" cellpadding="2" |
||
! Machine ||No. of Cores ||No. of Nodes ||Resolution ||Time for 1 Model Month |
! Machine ||No. of Cores ||No. of Nodes ||Resolution ||Time for 1 Model Month |
||
|- |
|- |
||
|HECToR Phase2b|| 96 || 4 || N48L60 || ~ |
|HECToR Phase2b|| 96 || 4 || N48L60 || ~46 minutes |
||
|- |
|- |
||
|MONSooN || 64 || 2 || N48L60 || ~ |
|MONSooN Phase 1|| 64 || 2 || N48L60 || ~51 minutes |
||
|- |
|||
|ARCHER|| 72 || 3 || N48L60 || ~25 minutes |
|||
|- |
|- |
||
|} |
|} |
||
== |
==Simple Tropospheric Chemistry with GLOMAP-mode aerosol - ''UKCA-MODE''== |
||
:''See also [[Aerosol_Subproject|MODE Aerosol description page]].'' |
|||
The UKCA aerosol scheme 'GLOMAP-mode' (sometimes called UKCA-MODE when in the UM) is described in detail in [http://www.geosci-model-dev.net/3/519/2010/gmd-3-519-2010.html Mann et al ](2010) and simulates aerosol microphysics (nucleation, condensation,coagulation and cloud processing), dry deposition, sedimentation and in-cloud/below-cloud scavenging. The scheme simulates atmospheric aerosol in a size-resolved manner with the chemical and aerosol microphysical processes determining the evolution of the size and composition of several size modes. The scheme transports the mass of sulphate, sea-salt, black carbon, organic matter and dust, along with the particle number concentration in each size mode. The aerosol scheme is coded flexibly to allow the same code to run with several alternative aerosol configurations via FORTRAN-90 modules. |
|||
[[Image:O3_column.png|thumb|right|Ozone column in Dobson Units (DU) calculated from a UM7.3 N48L60 UKCA-StratChem run using interactive UKCA ozone in the UM radiation scheme. This is the mean of 21 model years representing 1990.]] This scheme using the FCM branch '''fcm:um_br/dev/luke/VN7.3_UKCA_CheM''' at '''revision 5703'''. It has 41 species (36 of which are tracers), 113 bimolecular reactions, 34 photolysis reactions (using rates determined using a 3D look-up table), 17 termolecular reactions and 5 heterogeneous reactions. 15 species are wet-deposited and 15 are dry-deposited (using a 2D dry-deposition scheme). A stratospheric <math>O_{3}</math> is standard (based on the budget of Lee ''et al'', ''JGR'' '''107''' [http://www.agu.org/journals/ABS/2002/2001JD000538.shtml DOI:10.1029/2001JD000538] (2002)), as are other diagnostics of interest, such as the <math>CH_{4}</math> lifetime and Stratosphere-Troposphere Exchange of <math>O_{3}</math>. |
|||
UKCA-MODE can be coupled to all of the various UKCA chemistry schemes, but in this standard job uses the simple tropospheric chemistry scheme, extended to treat the degradation of the aerosol precursor species: sulphur dioxide (SO2);dimethylsulphide (DMS); and a lumped monoterpene tracer. |
|||
The chemistry is integrated using the ASAD chemical package, using a sparse-matrix Newton-Raphson solver. While the UM dynamical timestep is every 20 model minutes, chemistry is called every model hour, although emissions are still performed every dynamical timestep. By default, lower boundary conditions set for the tracer concentrations of <math>N_{2}O</math>, <math>CH_{4}</math>, <math>H_{2}</math>, <math>CF_{2}Cl_{2}</math>, <math>CFCl_{3}</math>, <math>CH_{3}Br</math>, and <math>CH_{2}Br_{2}</math> are specified for the years 1950-2100. These values are also used in the UM radiation scheme, along with <math>CO_{2}</math> concentrations. The tracers are initialised to spun-up fields from a previous ''StratChem'' run, however it can take time for a Stratospheric run to equilibrate and spin-up times as long as 20 model years may be required if significant changes are made. |
|||
An interface between UKCA-MODE and the Edwards-Slingo radiation scheme allows the aerosol concentrations to interact with the radiation both directly (through scattering and absorption of incoming solar and outgoing long-wave radiation) and indirectly via modified cloud albedo. In the standard jobs, both these effects are computed in a passive/ diagnostic mode via use of a double-call to the radiation scheme. |
|||
The chemistry scheme used in these simulations is the similar to that described in [http://www.geosci-model-dev.net/2/43/2009/gmd-2-43-2009.html Morgenstern ''et al''], ''Geosci. Model Dev.'', '''2''', 43-57, (2009). |
|||
One of the standard jobs also includes [[Nudging|Nudging]], wherein the model dynamics variables are nudged or relaxed towards ECMWF Reanalysis data. This configuration is particularly useful for evaluating the MODE scheme against measurements. |
|||
===Timings=== |
===Timings=== |
||
When run with all the 31 aerosol tracers switched on, the UKCA-MODE code adds 12% to the cost of the Atmosphere model (see O'Connor et al, 2011 ICP report to DECC/DEFRA). Please also note that the MONSooN timings are for phase 1 - jobs will run more quickly on phase 2 and you should consider using more nodes than on phase 1. |
|||
It should be noted that due to "jitter" on the HPC platform, all timings are approximate. As a simulation progresses the time per model month may increase as the fields spin-up from their initial state. |
|||
{| border="1" |
{| border="1" cellpadding="2" |
||
! Machine ||No. of Cores ||No. of Nodes ||Resolution ||Time for 1 Model Month |
! Machine ||No. of Cores ||No. of Nodes ||Resolution ||Time for 1 Model Month |
||
|- |
|- |
||
| |
|MONSooN (non-nudged)|| 64 || 2 || N96L38 || ~2 hours |
||
|- |
|- |
||
|MONSooN || 64 || 2 || |
|MONSooN (nudged)|| 64 || 2 || N96L38 || ~2.5 hours |
||
|- |
|- |
||
|} |
|} |
||
| Line 242: | Line 134: | ||
==Useful Links== |
==Useful Links== |
||
* [[Restarting an Atmos-Ocean Integration]] |
|||
* [[Adding New UKCA Tracers]] |
|||
* [[Duplicating STASH between UM jobs]] |
|||
* [[Using the UMUI]] |
|||
*[http://ncas-cms.nerc.ac.uk/index.php NCAS-CMS] |
*[http://ncas-cms.nerc.ac.uk/index.php NCAS-CMS] |
||
*[http://subversion.apache.org/ Subversion (SVN)] |
*[http://subversion.apache.org/ Subversion (SVN)] |
||
| Line 251: | Line 147: | ||
*[http://ncas-cms.nerc.ac.uk//index.php?option=com_wrapper&Itemid=274 Xancil] |
*[http://ncas-cms.nerc.ac.uk//index.php?option=com_wrapper&Itemid=274 Xancil] |
||
*[http://www.atm.ch.cam.ac.uk/acmsu/asad/ ASAD] |
*[http://www.atm.ch.cam.ac.uk/acmsu/asad/ ASAD] |
||
[[Category:Support]] |
|||
Latest revision as of 18:45, 14 November 2018
This page contains documentation on UKCA standard jobs that have been released or will soon be released.
You may also wish to look at the bugfixes page.
You may also wish to look at the MONSooN IBM to Cray Transition page.
Please also see the Getting Started with UKCA page.
Standard Release Jobs
UKCA can be configured through the UMUI, via it's own configuration panel. This mostly controls all the options required to run UKCA, although some extra changes have been made with hand-edits. To make it easy to get started with UKCA, several standard jobs have been created at various resolutions. To get started with UKCA, it should be a simple matter to take a copy of the UKCA job that you wish to run, changing a few user-specific options, and submitting the job.
Details of the jobs on ARCHER can be found here.
Code Changes
To run a UKCA standard job, please take a copy of one of the jobs listed below. If you need to make code changes, you must make a new FCM branch, and merge in the UKCA branch which is used in the job that you are taking, then use your branch instead.
Please do not take a checkout of the UKCA branch used in the UMUI FCM panel (e.g. fcm:um_br/dev/luke/VN7.3_UKCA_CheM) and run from this as a working copy. If the fcm commit command is run from this directory it would commit any changes you have made to this branch. In this case, this would affect the branch for other UKCA users.
Job List
Available release jobs are:
| Release Job # | UM Version | UMUI Job-ID | Climate Model | Horizontal Resolution | Vertical Levels | Model Top | Scheme Used | FCM Branch | Revision Number | Documentation |
|---|---|---|---|---|---|---|---|---|---|---|
| RJ1.0 | 7.3 | HECToR:xfvfa,xfvfc MONSooN: xfvfb ARCHER: xfvfd |
QESM-A | N48 (2.5°×3.75°) | L60 | ~84km | Tropospheric Chemistry with Isoprene | fcm:um_br/dev/luke/VN7.3_UKCA_CheM | 13783 | TropIsop |
| 7.3 | HadGEM3-A r2.0 | N96 (1.25°×1.875°) | L63 | ~40km | Tropospheric Chemistry with Isoprene | |||||
| RJ2.0 | 7.3 | HECToR:xfvga,xfvgc MONSooN:xfvgb ARCHER: xfvgd |
QESM-A | N48 | L60 | ~84km | Stratospheric Chemistry | fcm:um_br/dev/luke/VN7.3_UKCA_CheM | 13783 | StratChem |
| 7.3 | HadGEM3-A r2.0 | N96 | L85 | 85km | Stratospheric Chemistry | |||||
| 7.3 | QESM-A | N48 | L60 | ~84km | Simple Tropospheric Chemistry with MODE Aerosol | |||||
| RJ3.0 | 7.3 | HECToR: MONSooN:xfxlb |
N96 | L38 | ~40km | Simple Tropospheric Chemistry with MODE Aerosol | fcm:um_br/dev/mdalvi/VN7.3_MODEBLN_Rad_Indir | 5953 | UKCA-MODE | |
| RJ3.1 | 7.3 | HECToR: MONSooN:xfxld |
N96 | L38 | ~40km | Simple Tropospheric Chemistry with MODE Aerosol (Nudged) | fcm:um_br/dev/mdalvi/VN7.3_MODEBLN_Rad_Indir fcm:um_br/dev/mdalvi/VN7.3_nudged_new | 5953 5586 |
UKCA-MODE | |
| RJ4.0 | 8.4 | ARCHER: xlavb MONSooN: xlava |
GA4.0 | N96 | L85 | 85km | CheST chemistry with GLOMAP-mode aerosols | fcm:um_br/pkg/Config/vn8.4_UKCA | 18311 | CheST+GLOMAP-mode |
| RJ5.0 | 11.0+ | See GA7.1 StratTrop suites table for nudged and free-running | GA7.1 | N96e | L85 | 85km | StratTrop (CheST) with GLOMAP-mode aerosol | Runs from the trunk with possible additional branches | Release Job UM11.0 |
Required UMUI Changes
Before submitting a UKCA job, you must first alter several options in the UMUI:
- User-id, Mail-id (email address), and Tic-Code:
Model Selection -> User Information and Target Machine -> General Details
The user-id is your username on the target machine (i.e. HECToR).
The tic-code will be the HECToR charging code, e.g. n02-chem. This code can be found by logging into the HECToR SAFE pages, and viewing your account information. - Target machine root extract directory (UM_ROUTDIR):
Model Selection -> FCM Configuration -> FCM Extract and Build directories and Output levels
This needs to be the full path to one of your directories on the target machine (HECToR/ARCHER /home directory or MONSooN /projects directory).
See also: Using the UMUI for information on Optional UMUI changes such as changing the run length, archiving, emissions etc
Tropospheric Chemistry with Isoprene - TropIsop
This scheme using the FCM branch fcm:um_br/dev/luke/VN7.3_UKCA_CheM at revision 13783 (updated). It has 56 species (49 of which are tracers), 115 bimolecular reactions, 35 photolysis reactions (using rates determined using a 2D look-up table) and 15 termolecular reactions. 23 species are wet-deposited and 32 are dry-deposited (using a 2D dry-deposition scheme). Tropospheric and budgets are outputted as standard, as are other diagnostics of interest, such as the lifetime and Stratosphere-Troposphere Exchange of .
The chemistry is integrated using the ASAD chemical package, using a sparse-matrix Newton-Raphson solver. While the UM dynamical timestep is every 20 model minutes, chemistry is called every model hour, although emissions are still performed every dynamical timestep. Top boundary conditions are used for and and is treated as a constant. The tracers are initialised to spun-up fields from a previous TropIsop run, however, we recommend that a 15 month-spin-up should be performed.
The chemistry scheme used in these simulations is the same as in Telford et al, Atmos. Chem. Phys., 10, 7117-7125, doi:10.5194/acp-10-7117-2010 (2010). A model description paper is also in preparation (O'Connor et al, 2011).
The tropospheric ozone concentrations in UKCA-TropIsop are biased slightly high. This is improved with using an interactive photolysis scheme, which will be released for general use soon.
Timings
It should be noted that due to "jitter" on the HPC platform, all timings are approximate. As a simulation progresses the time per model month may increase as the fields spin-up from their initial state. Please also note that the MONSooN timings are for phase 1 - jobs will run more quickly on phase 2 and you should consider using more nodes than on phase 1.
| Machine | No. of Cores | No. of Nodes | Resolution | Time for 1 Model Month |
|---|---|---|---|---|
| HECToR Phase2b | 96 | 4 | N48L60 | ~50 minutes |
| MONSooN Pahse 1 | 64 | 2 | N48L60 | ~59 minutes |
| ARCHER | 72 | 3 | N48L60 | ~27 minutes |
Stratospheric Chemistry - StratChem
- See also Stratospheric Chemistry description page.
This scheme using the FCM branch fcm:um_br/dev/luke/VN7.3_UKCA_CheM at revision 13783 (updated). It has 41 species (36 of which are tracers), 113 bimolecular reactions, 34 photolysis reactions (using rates determined using a 3D look-up table), 17 termolecular reactions and 5 heterogeneous reactions. 15 species are wet-deposited and 15 are dry-deposited (using a 2D dry-deposition scheme). A stratospheric is standard (based on the budget of Lee et al, JGR 107 DOI:10.1029/2001JD000538 (2002)), as are other diagnostics of interest, such as the lifetime and Stratosphere-Troposphere Exchange of .
The chemistry is integrated using the ASAD chemical package, using a sparse-matrix Newton-Raphson solver. While the UM dynamical timestep is every 20 model minutes, chemistry is called every model hour, although emissions are still performed every dynamical timestep. By default, lower boundary conditions set for the tracer concentrations of , , , , , , and are specified for the years 1950-2100. These values are also used in the UM radiation scheme, along with concentrations. The tracers are initialised to spun-up fields from a previous StratChem run, however it can take time for a Stratospheric run to equilibrate and spin-up times as long as 20 model years may be required if significant changes are made.
The chemistry scheme used in these simulations is the similar to that described in Morgenstern et al, Geosci. Model Dev., 2, 43-57, (2009).
Please also see the bugfixes page for further information on the initial conditions used in the release job.
Timings
It should be noted that due to "jitter" on the HPC platform, all timings are approximate. As a simulation progresses the time per model month may increase as the fields spin-up from their initial state. Please also note that the MONSooN timings are for phase 1 - jobs will run more quickly on phase 2 and you should consider using more nodes than on phase 1.
| Machine | No. of Cores | No. of Nodes | Resolution | Time for 1 Model Month |
|---|---|---|---|---|
| HECToR Phase2b | 96 | 4 | N48L60 | ~46 minutes |
| MONSooN Phase 1 | 64 | 2 | N48L60 | ~51 minutes |
| ARCHER | 72 | 3 | N48L60 | ~25 minutes |
Simple Tropospheric Chemistry with GLOMAP-mode aerosol - UKCA-MODE
- See also MODE Aerosol description page.
The UKCA aerosol scheme 'GLOMAP-mode' (sometimes called UKCA-MODE when in the UM) is described in detail in Mann et al (2010) and simulates aerosol microphysics (nucleation, condensation,coagulation and cloud processing), dry deposition, sedimentation and in-cloud/below-cloud scavenging. The scheme simulates atmospheric aerosol in a size-resolved manner with the chemical and aerosol microphysical processes determining the evolution of the size and composition of several size modes. The scheme transports the mass of sulphate, sea-salt, black carbon, organic matter and dust, along with the particle number concentration in each size mode. The aerosol scheme is coded flexibly to allow the same code to run with several alternative aerosol configurations via FORTRAN-90 modules.
UKCA-MODE can be coupled to all of the various UKCA chemistry schemes, but in this standard job uses the simple tropospheric chemistry scheme, extended to treat the degradation of the aerosol precursor species: sulphur dioxide (SO2);dimethylsulphide (DMS); and a lumped monoterpene tracer.
An interface between UKCA-MODE and the Edwards-Slingo radiation scheme allows the aerosol concentrations to interact with the radiation both directly (through scattering and absorption of incoming solar and outgoing long-wave radiation) and indirectly via modified cloud albedo. In the standard jobs, both these effects are computed in a passive/ diagnostic mode via use of a double-call to the radiation scheme.
One of the standard jobs also includes Nudging, wherein the model dynamics variables are nudged or relaxed towards ECMWF Reanalysis data. This configuration is particularly useful for evaluating the MODE scheme against measurements.
Timings
When run with all the 31 aerosol tracers switched on, the UKCA-MODE code adds 12% to the cost of the Atmosphere model (see O'Connor et al, 2011 ICP report to DECC/DEFRA). Please also note that the MONSooN timings are for phase 1 - jobs will run more quickly on phase 2 and you should consider using more nodes than on phase 1.
| Machine | No. of Cores | No. of Nodes | Resolution | Time for 1 Model Month |
|---|---|---|---|---|
| MONSooN (non-nudged) | 64 | 2 | N96L38 | ~2 hours |
| MONSooN (nudged) | 64 | 2 | N96L38 | ~2.5 hours |