Adding Emissions into a Tracer
At the end of the previous tutorial you will now know how to create new tracers for use by UKCA. However, after completing the tasks, your tracers will still be empty, as nothing has been put into those tracers. This tutorial will teach you how to create an emissions ancillary file that the UM will read, and that you can then tell UKCA to use and emit into your tracer(s).
Making a new Emissons Ancillary File
The UM uses its own format to read-in intial and emissions data, the ancillary file. UKCA makes use of these files for the surface and aircraft emissions, and these files can easily be made up from netCDF data using the Xancil program. However, before we can use Xancil to create our emissions ancillary, we may first need to use [http:/75/badc.nerc.ac.uk/help/software/xconv/ Xconv] to regrid the raw data to the correct resolution of the UM configuration that you are using.
Regridding data with Xconv
We are using a model at N96L85 resolution. In the horizonal this is 1.875 degrees by 1.25 degrees. There are 85 vertical levels.
The N96 UM grid
- Has 192 points in longitude (x) and 145 points in latitude (y)
- Starts at 0.0 longitude with a spacing of 1.875 degrees
- Starts at -90.0 latitude (i.e. the South Pole) with a spacing of 1.25 degrees
Horizontal Regridding
Horizontal regridding in Xconv is straight-forward. First, open your dataset in Xconv by
xconv -i file.nc
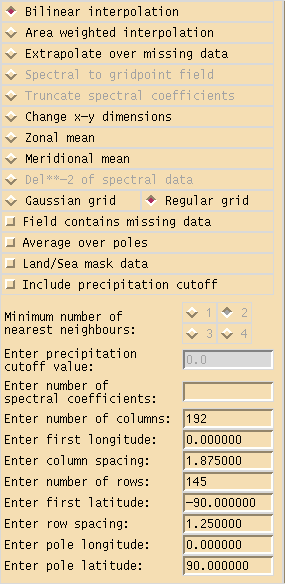
and double-click on the field that you want to regrid and then click the Trans button at the far top right. This will show a window similar to Figure 1 (which in fact shows the default settings for a N96 field).
As we are regridding an emission we need to select area weighted interpolation as we need to conserve the total amount of quantity emitted. Then scroll-down to the boxes at the end and enter the following values:
- Number of columns = 192
- First longitude = 0.000000
- Column spacing = 1.875000
- Number of rows = 145
- First latitude = -90.000000
- Row spacing = 1.250000
which match up with the grid definition above, and ensure that
- Pole longitude = 0.000000
- Pole latitude = 90.000000
Now click Apply. You should be given a message similar to
Area weighted interpolation from 720x360 Regular grid to 192x145 Regular grid
in the dialogue window. You can now extract this data as a new netCDF file (you cannot re-save a file in Xconv) by putting a name in the output file name box and clicking convert.
Vertical Regridding
It is not possible to vertically regrid data using Xconv. You will need to do this in another way. If you need to vertically regrid data, and am unsure of the best way, please contact Luke Abraham for advice.
Remember that
- Emissions data must be re-gridded in a mass conserving way, so you will probably need to integrate the field on one grid and then decompose it again on the new grid.
- Tracer data can be fitted to the profile of the field on the old grid.
Choosing a STASH slot for your new emission(s)
To make a new ancillary file for your new emission(s), you should first decide on a STASH item for it/them. Currently UKCA makes use of the user single-level ancillary file and user multi-level ancillary file which uses STASH section 0 items 301-320 (single-level) and 321-340 (multi-level). What these numbers correspond to is set in the file ukca_setd1defs.F90, as well as in the user STASHmaster file associated with the job you are using (which can be found in Model Selection → Atmosphere → STASH → User-STASHmaster files. Diags, Progs & Ancills).
Listing of emissions from STASH
| Stash code | Emission |
|---|---|
| 301 | NOx surf emissions |
| 302 | CH4 surf emissions |
| 303 | CO surf emissions |
| 304 | HCHO surf emissions |
| 305 | C2H6 surf emissions |
| 306 | C3H8 surf emissions |
| 307 | ME2CO surf emissions |
| 308 | MECHO surf emissions |
| 309 | C5H8 surf emissions |
| 310 | BC fossil fuel surf emissions |
| 311 | BC biofuel surf emissions |
| 312 | OC fossil fuel surf emissions |
| 313 | OC biofuel surf emissions |
| 314 | Monoterpene surf emissions |
| 315 | NVOC surf emissions |
| 322 | BC BIOMASS 3D EMISSION |
| 323 | OC BIOMASS 3D EMISSION |
| 340 | NOX AIRCRAFT EMS IN KG/S/GRIDCELL |
Code in ukca_setd1defs.F90
The species emitted are set in two places, firstly in the definition of an array called em_chem_spec which is scheme specific, and secondly in a block of code which searches through the em_chem_spec array and assigns a STASH number to it (as defined by the list above).
For the CheST/StratTrop chemistry (not using aerosol chemistry), em_chem_spec is set to
em_chem_spec = &
(/'NO ','CH4 ','CO ','HCHO ', &
'C2H6 ','C3H8 ','Me2CO ','MeCHO ', &
'C5H8 ','NO_aircrft'/)
This can be found in the Stratospheric Chemistry section, controlled by the IF block where (L_ukca_strattrop .AND. .NOT. L_ukca_achem). If you are using aerosol chemistry then the number of emissions are increased accordingly.
Further down the code there is this block of code:
J = n_use_tracers
IF (n_chem_emissions+n_3d_emissions+n_mode_emissions > 0) THEN
DO i=1,n_chem_emissions + n_3d_emissions
UkcaD1Codes(J+i)%section = 0
UkcaD1Codes(J+i)%item = n_emiss_first+i-1 ! trop chemistry
UkcaD1Codes(J+i)%len_dim1 = row_length ! uses stash codes
UkcaD1Codes(J+i)%len_dim2 = rows ! 301-309 for
UkcaD1Codes(J+i)%required = .true. ! surface emissions
UkcaD1Codes(J+i)%prognostic = .true. ! from Section 0
! Special cases, emissions already available in UM
IF (em_chem_spec(i)(1:7) == 'SO2_low') THEN
UkcaD1Codes(J+i)%item = 58
IF (.NOT. L_SO2_SURFEM .AND. (L_ukca_aerchem .OR. &
L_ukca_achem)) THEN
cmessage='SO2 surface emissions from UM are not flagged'
errcode=58
CALL EREPORT('UKCA_SETD1DEFS',errcode,cmessage)
ENDIF
ELSEIF (em_chem_spec(i)(1:7) == 'SO2_nat') THEN
UkcaD1Codes(J+i)%item = 121
UkcaD1Codes(J+i)%len_dim3 = tr_levels
IF (.NOT. L_SO2_NATEM .AND. (L_ukca_aerchem .OR. &
L_ukca_achem)) THEN
cmessage='SO2 natural emissions from UM are not flagged'
errcode=121
CALL EREPORT('UKCA_SETD1DEFS',errcode,cmessage)
ENDIF
ELSEIF (em_chem_spec(i)(1:8) == 'SO2_high') THEN
UkcaD1Codes(J+i)%item = 126
IF (.NOT. L_SO2_HILEM .AND. (L_ukca_aerchem .OR. &
L_ukca_achem)) THEN
cmessage='SO2 high-level emissions are not flagged'
errcode = UkcaD1Codes(J+i)%item
CALL EREPORT('UKCA_SETD1DEFS',errcode,cmessage)
ENDIF
ELSEIF (em_chem_spec(i)(1:3) == 'NH3') THEN
UkcaD1Codes(J+i)%item = 127
IF (.NOT. L_NH3_EM .AND. (L_ukca_aerchem .OR. &
L_ukca_achem)) THEN
cmessage='NH3 surface emissions from UM are not flagged'
errcode = UkcaD1Codes(J+i)%item
CALL EREPORT('UKCA_SETD1DEFS',errcode,cmessage)
ENDIF
ELSEIF (em_chem_spec(i) == 'BC_fossil ') THEN
UkcaD1Codes(J+i)%item = 310
ELSEIF (em_chem_spec(i) == 'BC_biofuel') THEN
UkcaD1Codes(J+i)%item = 311
ELSEIF (em_chem_spec(i) == 'OC_fossil ') THEN
UkcaD1Codes(J+i)%item = 312
ELSEIF (em_chem_spec(i) == 'OC_biofuel') THEN
UkcaD1Codes(J+i)%item = 313
ELSEIF (em_chem_spec(i) == 'Monoterp ') THEN
UkcaD1Codes(J+i)%item = 314
ELSEIF (em_chem_spec(i) == 'NVOC ') THEN
UkcaD1Codes(J+i)%item = 315
ELSEIF (em_chem_spec(i) == 'BC_biomass') THEN
UkcaD1Codes(J+i)%item = 322
UkcaD1Codes(J+i)%len_dim3 = tr_levels
ELSEIF (em_chem_spec(i) == 'OC_biomass') THEN
UkcaD1Codes(J+i)%item = 323
UkcaD1Codes(J+i)%len_dim3 = tr_levels
ELSEIF (em_chem_spec(i) == 'SO2_biomas') THEN
UkcaD1Codes(J+i)%item = 324
UkcaD1Codes(J+i)%len_dim3 = tr_levels
ELSEIF (em_chem_spec(i)(1:3) == 'DMS') THEN
UkcaD1Codes(J+i)%section = 17
UkcaD1Codes(J+i)%item = 205
UkcaD1Codes(J+i)%prognostic = .false.
IF (.NOT. L_DMS_EM .AND. (L_ukca_aerchem .OR. &
L_ukca_achem)) THEN
cmessage='DMS surface emissions from UM are not flagged'
errcode = UkcaD1Codes(J+i)%section*1000 + &
UkcaD1Codes(J+i)%item
CALL EREPORT('UKCA_SETD1DEFS',errcode,cmessage)
ENDIF
ELSEIF (em_chem_spec(i)(1:7) == 'NO_airc') THEN
UkcaD1Codes(J+i)%item = 340
UkcaD1Codes(J+i)%len_dim3 = tr_levels
ENDIF
ENDDO
ENDIF
This block of code is rather complicated, but what it essentially means is that for the STASH codes 301-309, the emissions are assumed to be in the order of the species in em_chem_spec, but for the other emissions the STASH numbers are explicitly defined. As you can see from the table above, for 2D (surface) emissions the numbers 301-315 are reserved, and for 3D emissions the numbers 322, 323, and 340 are reserved.
This means that if you are adding in a new surface emission(s) it is best to use the slots 316-320, unless you need more than one slot. For 3D emissions you have more leeway.
Emissions STASHmaster File
Now that you have selected your slot(s), you need to create a new STASH specification for it/them. The easiest way to do this is to copy the existing user STASHmaster file that defines your current (possible) emissions, and extend that. This is found in Model Selection → Atmosphere → STASH → User-STASHmaster files. Diags, Progs & Ancills. This contains entries like
# 1| 1 | 0 | 301 |NOx surf emissions | 2| 2 | 0 | 1 | 1 | 5 | -1 | -1 | 0 | 0 | 0 | 0 | 3| 000000000000000000000000000000 | 00000000000000000001 | 3 | 4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | 5| 0 | 531 | 0 | 129 | 0 | 0 | 0 | 0 | 0 | # # 1| 1 | 0 | 340 |NOX AIRCRAFT EMS IN KG/S/GRIDCELL | 2| 2 | 0 | 1 | 1 | 2 | 10 | 11 | 0 | 0 | 0 | 0 | 3| 000000000000000000000000000000 | 00000000000000000001 | 3 | 4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | 5| 0 | 520 | 20 | 65 | 0 | 0 | 0 | 9999 | 0 | #
The top entry (NOx surf emissions) defines a 2D field, and the other (NOX AIRCRAFT EMS IN KG/S/GRIDCELL) defines a 3D field. You can see that there are differences in the numbers (other than the 301/340 item number) in various places in these specifications, which effectively (in this instance) tell STASH if the field is 2D or 3D.
Full details on what each of these numbers mean can be found in appendix 3 of Unified Model Documentation Paper C4 which can be found on the Met Office Collaboration Twiki (password required).
You should copy either the 2D or 3D specification, depending on what type of emission you are adding in, and edit only the STASH item number, name of field, and the field code. These can be found here:
# 1| 1 | 0 | 301 |NOx surf emissions | 2| 2 | 0 | 1 | 1 | 5 | -1 | -1 | 0 | 0 | 0 | 0 | 3| 000000000000000000000000000000 | 00000000000000000001 | 3 | 4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | 5| 0 | 531 | 0 | 129 | 0 | 0 | 0 | 0 | 0 | #
For the field code (531 above), if you were making a new emission at 316, it is advised that you increase the current code above by 15 as well, e.g. 546.
You should make these changes to your copy of the original emissions user STASHmaster file, save this, and then replace the original file in the UMUI's Model Selection → Atmosphere → STASH → User-STASHmaster files. Diags, Progs & Ancills table with your new file. Now go to Model Selection → Atmosphere → STASH → Initialisation of User Prognostics and scroll down the table until you find your new emission. Set the value in the Option column to 2 (Initialise to User Ancillary File).
As you need to make up a new ancillary file, you should copy this user STASHmaster file onto the supercomputer, as it will be needed by Xancil when you make the new ancillary file. It is advisable to put it in the same directory as the one contaning your new emission(s) field.
Using Xancil
Extract your current emissions
Due to current limitations of the UM, you can only have one user single-level and one multi-level file. If you want to add a new emissions field, you must include the existing emissions in your new file along with it.
You can use Xconv to extract these fields. You must first find the location of the current file(s). To do this, go to Model Selection → Atmosphere → Ancillary data and input data files → Climatologies and potential climatologies and either open the User multi-level ancillary file & fields or User single-level ancillary file & fields panel. This will give two boxes giving the file name and the directory name or environment variable. This second box will using contain an environment variable which sets the directory location. You can find the value of this environment variable in Model Selection → Input/Output Control and Resources → Time Convention and SCRIPT Environment Variables.
Once you have found the required file, extract all the fields within it to one of your own directories (ideally the one containing the netCDF file of your new emission(s) field).
Make a new emissions ancillary file
Xancil is installed on both MONSooN (on the postproc03 machine) at
/projects/um1/linux/bin/xancil
and on HECToR at
/work/n02/n02/hum/bin/xancil
You may already have this location in your PATH and so can just launch Xancil from the command line by typing xancil. When you do this it will load up the Xancil window, which is initially rather empty. You should click on the Xancil text in the top-left corner. This will give 4 options:
- Configuration
- Atmosphere Ancillary Files
- Ocean Ancillary Files
- Generalised Ancillary Files
When using UKCA you will need to make use of all of these options, with the exception of the Ocean Ancillary Files.
You should view the Using Xancil page for more details on each of these sub-menus.
Load your netCDF and user STASHmaster files into the Xancil → Configuration panel, and define the vertical levels if you are making a 3D ancillary file.
Click on the Atmosphere Ancillary Files text and either open the Multi-level User Fields or Single-level User Fields panels. You should now
- Set the output file name
- Define the dates. If the ones in the netCDf file are fine to use then you can use them, or you can specify them. For this latter option you can either use the dates from the grid configuration panel, or you can define them again here
- Enter the number of ancillary fields. This will need to be the total of the number of fields in the original ancillary file, plus the number of new emissions you are adding
- For each individual field you should select it with the mouse, then
- Define which netCDF file to use (it will remember the preference from the previous field)
- Enter the STASH code (the PP code should be filled in automatically)
- Either enter or confirm the variable name. Xancil takes the variable name from the field code defined in the STASHmaster file specification for each field. If there are multiple fields in the netCDF file with the same field code then Xancil may select the wrong one. If the name does not match a field code you will need to select it manually.
Once you have entered all the data for all fields you should
- Use the Save/Save As button to save the job, as it may be needed later
- Click the Output Anc. Files button to create the new ancillary file. Any errors will appear in the Output messages window, or to the terminal.
Written by Luke Abraham 2013