UKCA Chemistry and Aerosol vn10.9 Tutorial 10: Difference between revisions
No edit summary |
|||
| (75 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
==What you will learn in this Tutorial== |
==What you will learn in this Tutorial== |
||
In this tutorial you will learn how to output and process aerosol diagnostics from UKCA. |
|||
===Python=== |
|||
In this Tutorial you will make extensive use of Python to process the UM output. Instructions as to how to use this are in [[UKCA_Chemistry_and_Aerosol_vn10.9_Tutorial_5#Using_python_to_regrid_your_emissions|Tutorial 5]]. |
|||
Example python scripts are provided for each Task, and you should take the time to read through these and understand what they are doing. None of these scripts plot any of the output, so as an extension to these tasks you could try extending the scripts to plot the output from python directly, rather than visualising via Xconv. Further information (and examples) on how Iris plots can be found here: |
|||
* http://scitools.org.uk/iris/docs/latest/userguide/plotting_a_cube.html |
|||
* http://scitools.org.uk/iris/docs/latest/examples/General/global_map.html |
|||
==Task 10.1: Output aerosol diagnostics== |
==Task 10.1: Output aerosol diagnostics== |
||
<span style="color:green">'''TASK 10.1:''' Output the following aerosol and radiation diagnostics to the '''UPA''' output stream. You should make a new ''time profile'' (called TRAD) to only output these on '''radiation timesteps'''. You will also need to make a new ''domain profile'' (called D3DAR) for s02i530 and s02i540.</span> |
<span style="color:green">'''TASK 10.1:''' Output the following aerosol and radiation diagnostics to the '''UPA''' output stream. You should make a new ''time profile'' (called TRAD) to only output these on '''radiation timesteps'''. You will also need to make a new ''domain profile'' (called D3DAR) for s02i530 and s02i540, to '''output these on both model levels and pseudo levels'''.</span> |
||
{| class="collapsible collapsed wikitable" |
{| class="collapsible collapsed wikitable" |
||
| Line 12: | Line 22: | ||
! Hint |
! Hint |
||
|- |
|- |
||
| Remember to output your aerosol diagnostics on the AOT pseudo levels using the DIAGAOT profile. |
|||
| You can find the number of radiation timesteps in a 24-hour period at: um <math>\rightarrow</math> namelist <math>\rightarrow</math> UM Science Settings <math>\rightarrow</math> Section 01 - 02 Radiation <math>\rightarrow</math> Shortwave. |
|||
|} |
|} |
||
| Line 58: | Line 68: | ||
| 2 || 540 || CLASSIC 3D AEROSOL EXTINCTION |
| 2 || 540 || CLASSIC 3D AEROSOL EXTINCTION |
||
|} |
|} |
||
==Radiation Timesteps== |
|||
The number of shortwave radiation timesteps per day is given by '''<code>i_sw_radstep_perday_prog</code>''', found in um <math>\rightarrow</math> namelist <math>\rightarrow</math> UM Science Settings <math>\rightarrow</math> Section 01 - 02 Radiation <math>\rightarrow</math> Shortwave. The equivalent for longwave is '''<code>i_lw_radstep_perday_prog</code>''', found in um <math>\rightarrow</math> namelist <math>\rightarrow</math> UM Science Settings <math>\rightarrow</math> Section 01 - 02 Radiation <math>\rightarrow</math> Longwave. |
|||
In the UKCA training suite, both of these are set to '''16''', i.e. a call every 90 minutes. This value is very configuration dependent - sometimes radiation is called every hour (24), sometimes every 3 hours (8) etc. You should check the settings in your suite if you need to consider any of the radiation diagnostics. |
|||
==Making new STASH profiles== |
|||
While we have covered outputting and creating new diagnostics in [[UKCA_Chemistry_and_Aerosol_vn10.9_Tutorial_3|Tutorial 3]], [[UKCA_Chemistry_and_Aerosol_vn10.9_Tutorial_4|Tutorial 4]], [[UKCA_Chemistry_and_Aerosol_vn10.9_Tutorial_5|Tutorial 5]], and [[UKCA_Chemistry_and_Aerosol_vn10.9_Tutorial_9|Tutorial 9]], you were only making use of existing profiles. Here you will learn how to make new '''time''', '''domain''', and '''usage''' profiles. |
|||
===Time profiles=== |
|||
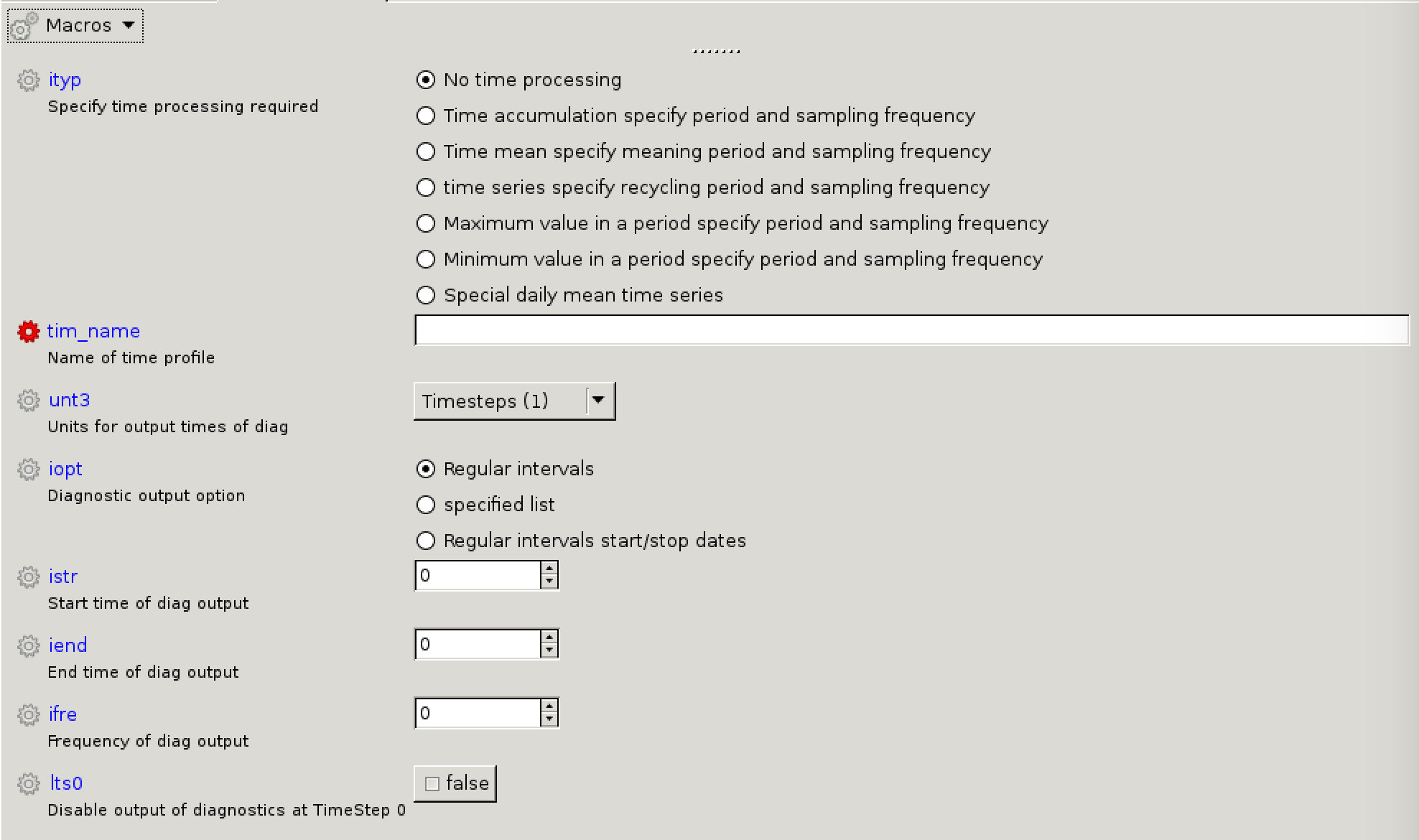
[[Image:Vn10.9_newtime.png|thumb|300px|right|The new time panel in Rose.]] |
|||
To make a new time profile, go to: '''um <math>\rightarrow</math> namelist <math>\rightarrow</math> Model Input and Output <math>\rightarrow</math> STASH Requests and Profiles <math>\rightarrow</math> Time Profiles'''. This contains a list of existing time profiles, with names like ''<code>t3hmn_039ecafe</code>'' etc. |
|||
To make a new time profile, you should: |
|||
# Right-click anywhere on the list and click the blue plus symbol ('''<span style="color:blue">+</span>''') named '''new section''' |
|||
#: This will make a new blank line (usually labelled '''1''', the next '''2''' etc.) with a red '''<span style="color: red">X</span>''' next to it. |
|||
# Right-click this new line and click '''View namelist'''. |
|||
# Fill-in the values as required for what you want to do. |
|||
# When you have finished, you will need to run the When you have finished, you will need to run the '''stashindices.TidyStashTransform''' macro by going to the '''Metadata <math>\rightarrow</math> um''' drop-down menu. This will rename the new profile into the correct format. |
|||
====Example: Output on Radiation Timesteps==== |
|||
You will have discovered that there are 16 radiation timesteps per day, but to work out how to output diagnostics you need to know many model timesteps per day there are. This can be found at: um <math>\rightarrow</math> namelist <math>\rightarrow</math> Top Level Model Control <math>\rightarrow</math> Model Domain and Timestep. The number is given by the variable '''<code>steps_per_periodim</code>''', which is defined as the number of steps per period given (in seconds) by '''<code>secs_per_periodim</code>'''. For one day, this value is 86400. |
|||
In the UKCA training suite, <code>secs_per_periodim</code>=86400, and <code>steps_per_periodim</code>=48. This means that there are 48 timesteps per day, i.e. each timestep is '''30 minutes'''. |
|||
We can combine this with the number of radiation timesteps per day (16), to calculate that '''radiation is called every third model timestep''' (48/16 = 3) in this configuration. We can now fill-in the values in the new time profile accordingly, e.g. |
|||
* '''<code>ityp</code>:''' No time processing |
|||
* '''<code>tim_name</code>:''' e.g. TRAD |
|||
* '''<code>unt3</code>:''' Timesteps (1) |
|||
* '''<code>iopt</code>:''' Regular intervals |
|||
* '''<code>istr</code>:''' 1 (i.e. start on first timestep) |
|||
* '''<code>iend</code>:''' -1 (i.e. never stop outputting) |
|||
* '''<code>ifre</code>:''' 3 (i.e. output every third timestep) |
|||
We need to start at timestep 1, as radiation is called on the first timestep, and then called every third model timestep after that. In contrast, the UKCA Newton-Raphson chemical solver is first called on the first hour, and then every hour after that. For the UKCA training suite this would mean that <code>istr</code>=2 and <code>ifre</code>=2. |
|||
===Domain profiles=== |
|||
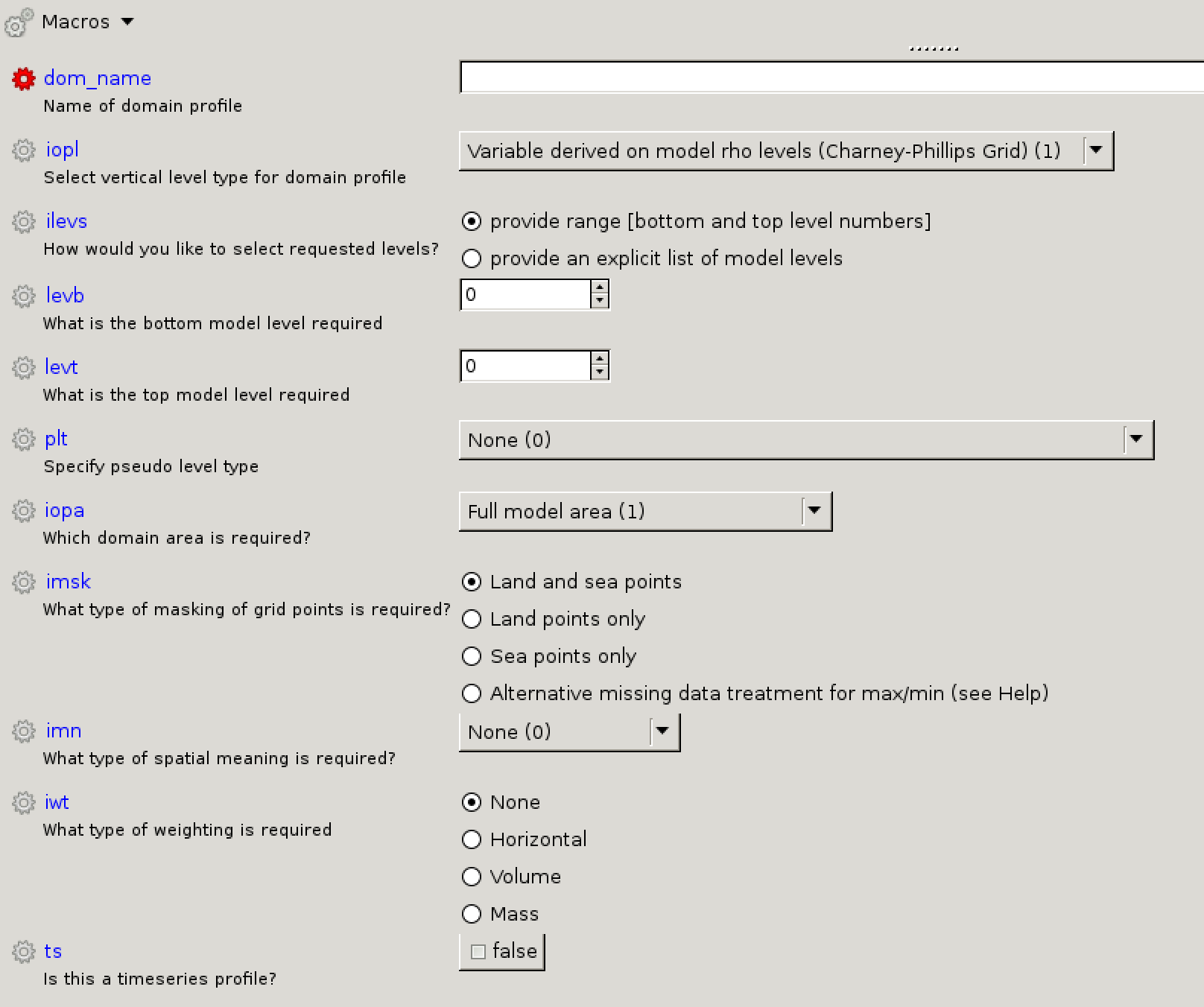
[[Image:Vn10.9_newdom.png|thumb|300px|right|The new domain panel in Rose.]] |
|||
To make a new time profile, go to: '''um <math>\rightarrow</math> namelist <math>\rightarrow</math> Model Input and Output <math>\rightarrow</math> STASH Requests and Profiles <math>\rightarrow</math> Domain Profiles'''. This contains a list of existing domain profiles, with names like ''<code>dallth_1e2e730d</code>'' etc. |
|||
To make a new domain profile, you should: |
|||
# Right-click anywhere on the list and click the blue plus symbol ('''<span style="color:blue">+</span>''') named '''new section''' |
|||
#: This will make a new blank line (usually labelled '''1''', the next '''2''' etc.) with a red '''<span style="color: red">X</span>''' next to it. |
|||
# Right-click this new line and click '''View namelist'''. |
|||
# Fill-in the values as required for what you want to do. |
|||
# When you have finished, you will need to run the When you have finished, you will need to run the '''stashindices.TidyStashTransform''' macro by going to the '''Metadata <math>\rightarrow</math> um''' drop-down menu. This will rename the new profile into the correct format. |
|||
====Example: Output AOT pseudo levels on all model theta levels==== |
|||
Aerosol diagnostics are defined on a series of 6 '''pseudo levels''', corresponding to 0.38, 0.44, 0.55, 0.67, 0.87, and 1.02 microns. You will therefore need to output on your data on all of these. For some diagnostics, such as AOD, these are defined on a single level, but others, such as extinction, are a 3D field over the whole atmosphere. Therefore, to output the 3D aerosol extinction we need to make a new profile over all model (theta grid) levels, ''and'' all 6 pseudo levels. |
|||
We can now fill-in the values in the new domain profile accordingly, e.g. |
|||
* '''<code>dom_name</code>:''' e.g. D3DAR |
|||
* '''<code>iopl</code>:''' Variable derived on model theta levels (Charney-Phillips Grid) (2) |
|||
* '''<code>ilevs</code>:''' provide range <nowiki>[</nowiki>bottom and top level numbers<nowiki>]</nowiki> |
|||
* '''<code>ilevb</code>:''' 1 (i.e. 20m level) |
|||
* '''<code>ilevt</code>:''' 85 (i.e. highest level) |
|||
* '''<code>plt</code>:''' Radiation bands for calculating aerosol optical depth (4) |
|||
: This will then insert a new option below (<code>pslist</code>) to allow you to specify the bands you require. |
|||
* '''<code>pslist</code>:''' press the blue plus symbol ('''<span style="color:blue">+</span>''') 5 times, and enter the numbers 1 to 6 in each of the boxes to output all AOT pseudo levels. |
|||
* '''<code>iopa</code>:''' Full model area (1) |
|||
* '''<code>imsk</code>:''' Land and sea points |
|||
* '''<code>imn</code>:''' None (0) |
|||
* '''<code>iwt</code>:''' None |
|||
* '''<code>ts</code>:''' false |
|||
===Usage profiles=== |
|||
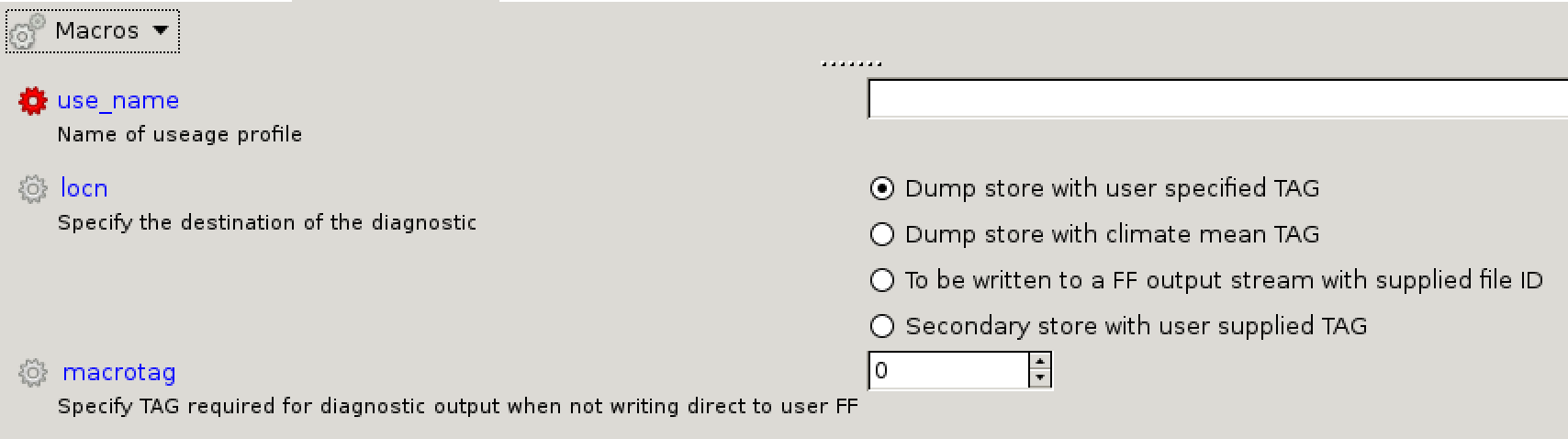
[[Image:Vn10.9_newuse.png|thumb|300px|right|The new usage panel in Rose.]] |
|||
Making new usage profiles is not as straight-forward as for time and domain profiles, as these are used to output diagnostics to different output files (and possibly to climate mean files). To see what output streams are available, go to: '''um <math>\rightarrow</math> namelist <math>\rightarrow</math> Model Input and Output <math>\rightarrow</math> Model Output Streams'''. In the UKCA training suite, only '''pp0''' is active. |
|||
To look at climate meaning, go to: '''um <math>\rightarrow</math> namelist <math>\rightarrow</math> Model Input and Output <math>\rightarrow</math> Dumping and Meaning'''. This is controlled by the logical '''<code>l_meaning_sequence</code>''', which in the UKCA training suite is set to '''false''' (i.e. no climate meaning). If this is set to ''true'' then various settings will need to be chosen, such as the reference time, the frequency of files, and which files to output. |
|||
To view the usage profiles, go to: '''um <math>\rightarrow</math> namelist <math>\rightarrow</math> Model Input and Output <math>\rightarrow</math> STASH Requests and Profiles <math>\rightarrow</math> Usage Profiles'''. This contains a list of existing domain profiles, with names like ''<code>upa_ffb3f00b</code>'' etc. If you look at the '''upa''' namelist (by right-clicking on it and clicking '''view namelist''') you can see that this corresponds with the ''pp0'' output stream. |
|||
It is also possible to write diagnostics to a '''tag''', which can then be read by other parts of the code. This is how UKCA couples to the UM, via tag=98. This can be seen in the '''upukca''' usage profile. |
|||
When you take a copy of an existing configuration it is unlikely that you will need to make any changes to any (or make new) usage profiles. However, if you do want to make a new usage profile, you should: |
|||
# Right-click anywhere on the list and click the blue plus symbol ('''<span style="color:blue">+</span>''') named '''new section''' |
|||
#: This will make a new blank line (usually labelled '''1''', the next '''2''' etc.) with a red '''<span style="color: red">X</span>''' next to it. |
|||
# Right-click this new line and click '''View namelist'''. |
|||
# Fill-in the values as required for what you want to do. |
|||
# When you have finished, you will need to run the When you have finished, you will need to run the '''stashindices.TidyStashTransform''' macro by going to the '''Metadata <math>\rightarrow</math> um''' drop-down menu. This will rename the new profile into the correct format. |
|||
'''Note''' that if you are making a new usage profile that is not associated with a ''tag'', you may also need to make changes to the output streams. This could be quite involved. |
|||
===Using your new profiles=== |
|||
Once you have made your new profiles you should then use them in the usual way when outputting diagnostics, as covered in [[UKCA_Chemistry_and_Aerosol_vn10.9_Tutorial_3#STASH_Panel|Tutorial 3]]. |
|||
When all your new STASH is set-up, you can use the Validator Macros to check if the names are correct, or if there are any unused STASH items etc. To run this, go to '''Metadata <math>\rightarrow</math> Check all Validator Macros''' from the drop-down menu. You can also check for ''fail-if'' or ''warn-if'' issues. However, these won't warn of incorrect domain requests (i.e. requesting pressure-levels for diagnostics only defined on a single level), so some care should still be taken. |
|||
==Solution to Task 10.1== |
==Solution to Task 10.1== |
||
| Line 63: | Line 174: | ||
You were given the task |
You were given the task |
||
* ''Output the following aerosol and radiation diagnostics to the '''UPA''' output stream. You should make a new ''time profile'' (called TRAD) to only output these on '''radiation timesteps'''. You will also need to make a new ''domain profile'' (called D3DAR) for s02i530 and s02i540.'' |
* ''Output the following aerosol and radiation diagnostics to the '''UPA''' output stream. You should make a new ''time profile'' (called TRAD) to only output these on '''radiation timesteps'''. You will also need to make a new ''domain profile'' (called D3DAR) for s02i530 and s02i540, to '''output these on both model levels and pseudo levels'''.'' |
||
{| class="wikitable" border = "1px" |
{| class="wikitable" border = "1px" |
||
| Line 111: | Line 222: | ||
and were given the hint |
and were given the hint |
||
:* ''Remember to output your aerosol diagnostics on the AOT pseudo levels using the DIAGAOT profile.'' |
|||
:* ''You can find the number of radiation timesteps in a 24-hour period at: um <math>\rightarrow</math> namelist <math>\rightarrow</math> UM Science Settings <math>\rightarrow</math> Section 01 - 02 Radiation <math>\rightarrow</math> Shortwave.'' |
|||
For a working Rose suite that has completed this task, please see |
For a working Rose suite that has completed this task, please see |
||
| Line 646: | Line 757: | ||
==Task 10.2: Calculate aerosol optical depth== |
==Task 10.2: Calculate aerosol optical depth== |
||
<span style="color:green">'''TASK 10.2:''' Calculate the aerosol optical depth at 0.55 microns.</span> |
<span style="color:green">'''TASK 10.2:''' Calculate the aerosol optical depth at 0.55 microns on the second radiation timestep.</span> |
||
{| class="collapsible collapsed wikitable" |
{| class="collapsible collapsed wikitable" |
||
| Line 652: | Line 763: | ||
! Hint |
! Hint |
||
|- |
|- |
||
| The UM |
| The UM calculates optical depth diagnostics on 6 pseudo levels corresponding to 0.38, 0.44, 0.55, 0.67, 0.87, and 1.02 microns, therefore 0.55 microns is pseudo level 3. |
||
|} |
|} |
||
| Line 719: | Line 830: | ||
# output to netCDF |
# output to netCDF |
||
iris.save(aodsum,'Task102_AOD.nc',netcdf_format='NETCDF3_CLASSIC') |
iris.save(aodsum,'Task102_AOD.nc',netcdf_format='NETCDF3_CLASSIC') |
||
You should edit this script to point to your UM output file, and will need to |
|||
module load anaconda/2.2.0-python2 |
|||
before running it, e.g. |
|||
python -c 'execfile("write_AOD.py")' |
|||
==Solution to Task 10.2 == |
==Solution to Task 10.2 == |
||
| Line 733: | Line 836: | ||
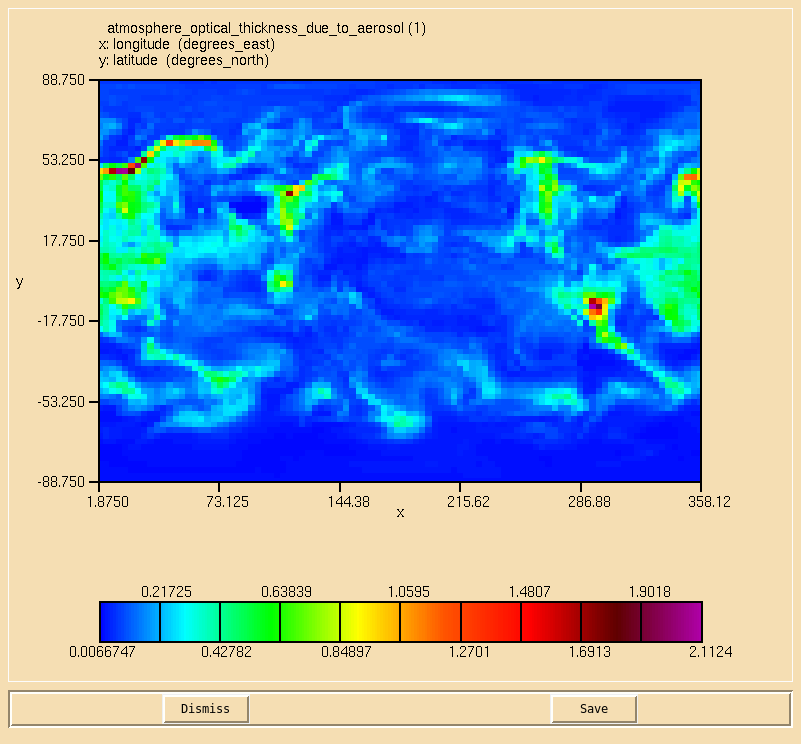
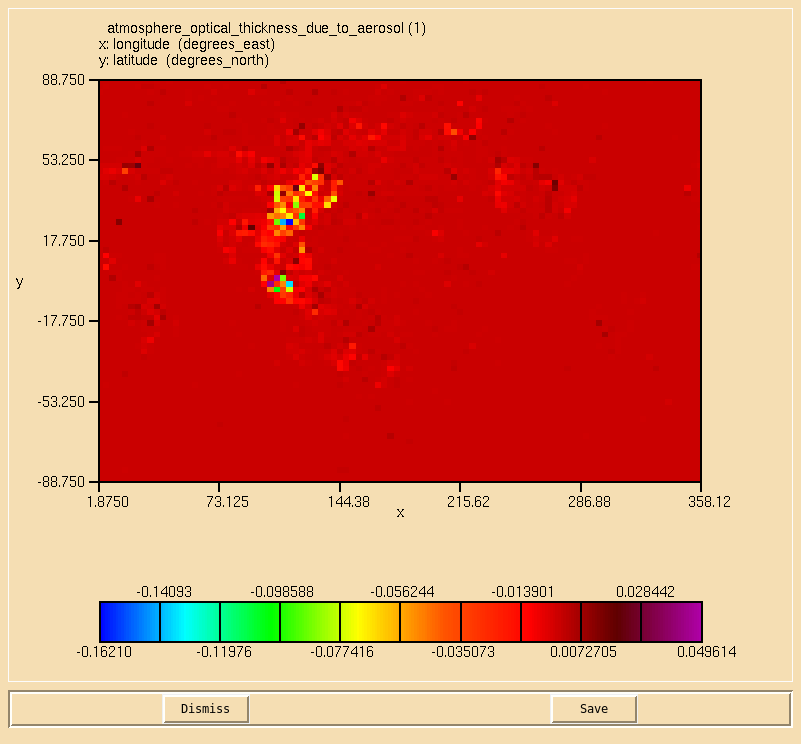
Image:Vn10.9_102_AOD.png|0.55 micron AOD calculated from the component aerosol AOD diagnostics. |
Image:Vn10.9_102_AOD.png|0.55 micron AOD calculated from the component aerosol AOD diagnostics. |
||
</gallery> |
</gallery> |
||
You were asked to |
|||
* ''Calculate the aerosol optical depth at 0.55 microns on the second radiation timestep.'' |
|||
and were given the hint |
|||
:* ''The UM calculates optical depth diagnostics on 6 pseudo levels corresponding to 0.38, 0.44, 0.55, 0.67, 0.87, and 1.02 microns, therefore 0.55 microns is pseudo level 3.'' |
|||
You should make use of the python script provided to do this. |
|||
Sample output from this task can be found in the '''<code>/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.2/</code>''' directory on ARCHER, containing the following: |
Sample output from this task can be found in the '''<code>/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.2/</code>''' directory on ARCHER, containing the following: |
||
| Line 740: | Line 853: | ||
==Task 10.3: Calculate the single-scattering albedo== |
==Task 10.3: Calculate the single-scattering albedo== |
||
<span style="color:green">'''TASK 10.3:''' Calculate the single-scattering albedo at 0.55 microns, defined as: </span> |
<span style="color:green">'''TASK 10.3:''' Calculate the single-scattering albedo at 0.55 microns on the second radiation timestep, defined as: </span> |
||
<math> |
<math> |
||
| Line 826: | Line 939: | ||
==Solution to Task 10.3 == |
==Solution to Task 10.3 == |
||
<gallery widths=300px heights=300px> |
|||
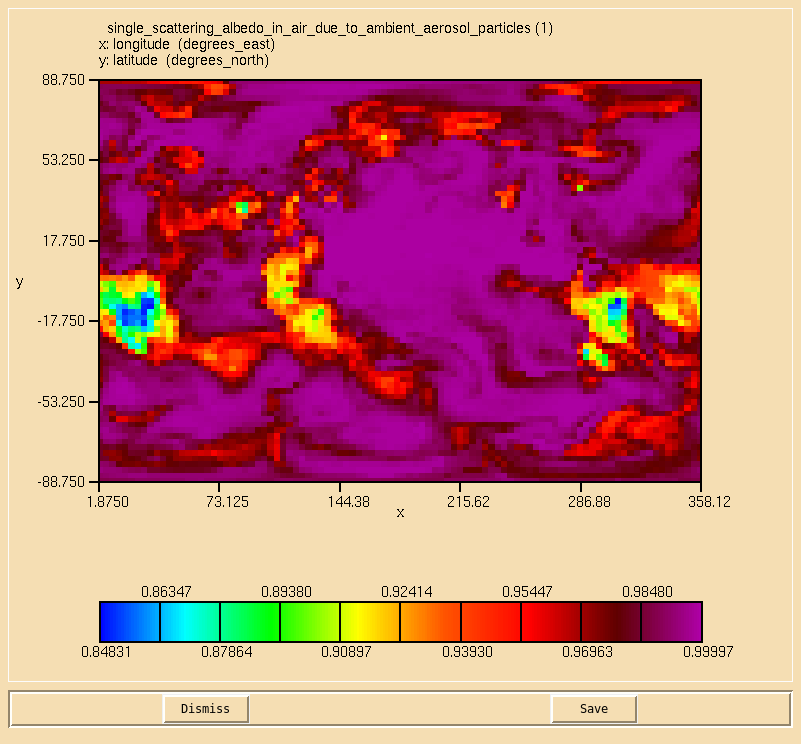
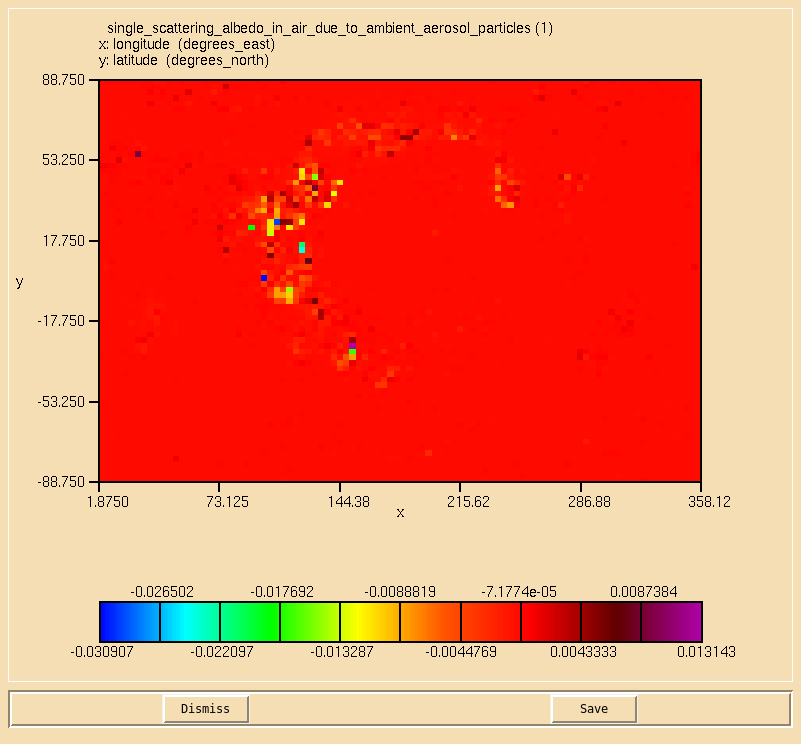
Image:Vn10.9_103_SSA.png|0.55 micron single-scattering albedo. |
|||
</gallery> |
|||
You were asked to |
|||
* ''Calculate the single-scattering albedo at 0.55 microns on the second radiation timestep, defined as:'' |
|||
<math> |
|||
1 - \left(\frac{\textrm{Absorption\ Aerosol\ Optical\ Depth}}{\textrm{Aerosol\ Optical\ Depth}}\right) |
|||
</math> |
|||
You should use the python script provided to do this. |
|||
Sample output from this task can be found in the '''<code>/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.3/</code>''' directory on ARCHER, containing the following: |
Sample output from this task can be found in the '''<code>/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.3/</code>''' directory on ARCHER, containing the following: |
||
| Line 833: | Line 960: | ||
==Task 10.4: Calculate the top of the atmosphere net downward radiative flux== |
==Task 10.4: Calculate the top of the atmosphere net downward radiative flux== |
||
<span style="color:green">'''TASK 10.4:''' </span> |
<span style="color:green">'''TASK 10.4:''' Calculate the net downward top of the atmosphere radiative flux on the second radiation timestep.</span> |
||
===Python script=== |
|||
To calculate the net TOA downward radiative flux, you should sum up the outgoing contributions from shortwave and longwave radiation, and take this away from the incoming shortwave radiative flux. |
|||
On ARCHER, an example python script to do this has been provided at '''<code>/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.4/write_TOA.py</code>''': |
|||
#!/usr/bin/env python |
|||
# This file is part of the UKCA Tutorials: |
|||
# http://www.ukca.ac.uk/wiki/index.php/UKCA_Chemistry_and_Aerosol_Tutorials_at_vn10.9 |
|||
# Copyright (C) 2017 University of Cambridge |
|||
# This is free software: you can redistribute it and/or modify it under the |
|||
# terms of the GNU Lesser General Public License as published by the Free |
|||
# Software Foundation, either version 3 of the License, or (at your option) |
|||
# any later version. |
|||
# It is distributed in the hope that it will be useful, but WITHOUT ANY |
|||
# WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR |
|||
# A PARTICULAR PURPOSE. See the GNU Lesser General Public License for more |
|||
# details. |
|||
# You find a copy of the GNU Lesser General Public License at <http://www.gnu.org/licenses/>. |
|||
# Written by N. Luke Abraham 2017-12-11 <nla27@cam.ac.uk> |
|||
# preamble |
|||
import iris |
|||
import iris.time |
|||
fname='/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.1/atmosa.pa19810901_00' |
|||
# constraint on time to get 2nd radiation timestep |
|||
tconstr=iris.Constraint(time=lambda cell: cell.point.hour == 2) |
|||
# load all TOA components at 0.55 micron |
|||
# must use this way of loading to account for constraint on time |
|||
with iris.FUTURE.context(cell_datetime_objects=True): |
|||
isw=iris.load_cube(fname,[iris.AttributeConstraint(STASH='m01s01i207') & tconstr]) |
|||
osw=iris.load_cube(fname,[iris.AttributeConstraint(STASH='m01s01i208') & tconstr]) |
|||
olw=iris.load_cube(fname,[iris.AttributeConstraint(STASH='m01s02i205') & tconstr]) |
|||
# make cube to store net downward TOA flux |
|||
toa=isw.copy() |
|||
# add-up components |
|||
toa.data=isw.data - (osw.data + olw.data) |
|||
toa.rename('toa_net_downward_radiative_flux') |
|||
# remove unlimited dimension when writing to netCDF |
|||
iris.FUTURE.netcdf_no_unlimited=True |
|||
# output to netCDF |
|||
iris.save(toa,'Task104_TOA.nc',netcdf_format='NETCDF3_CLASSIC') |
|||
==Solution to Task 10.4 == |
==Solution to Task 10.4 == |
||
<gallery widths=300px heights=300px> |
|||
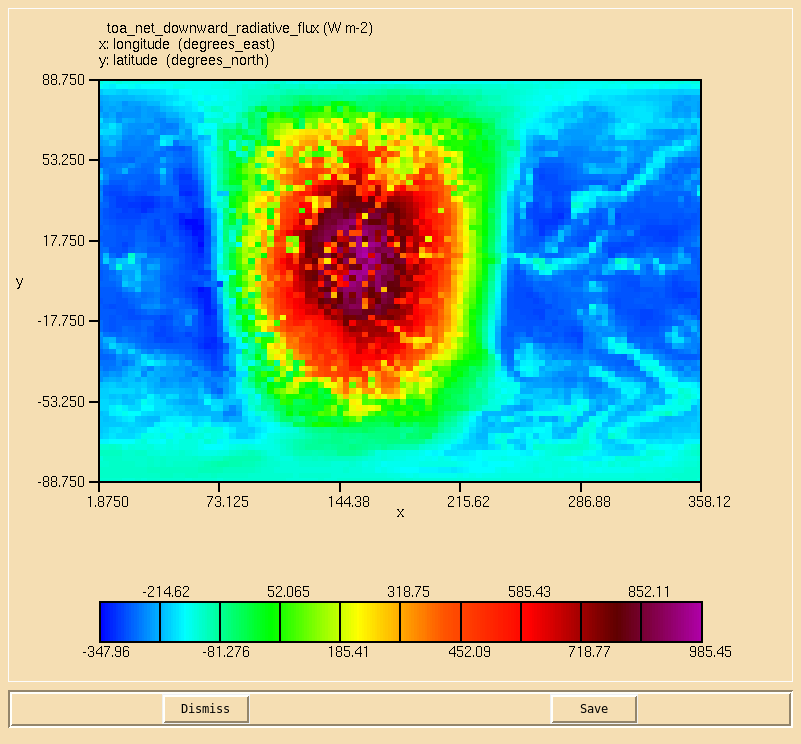
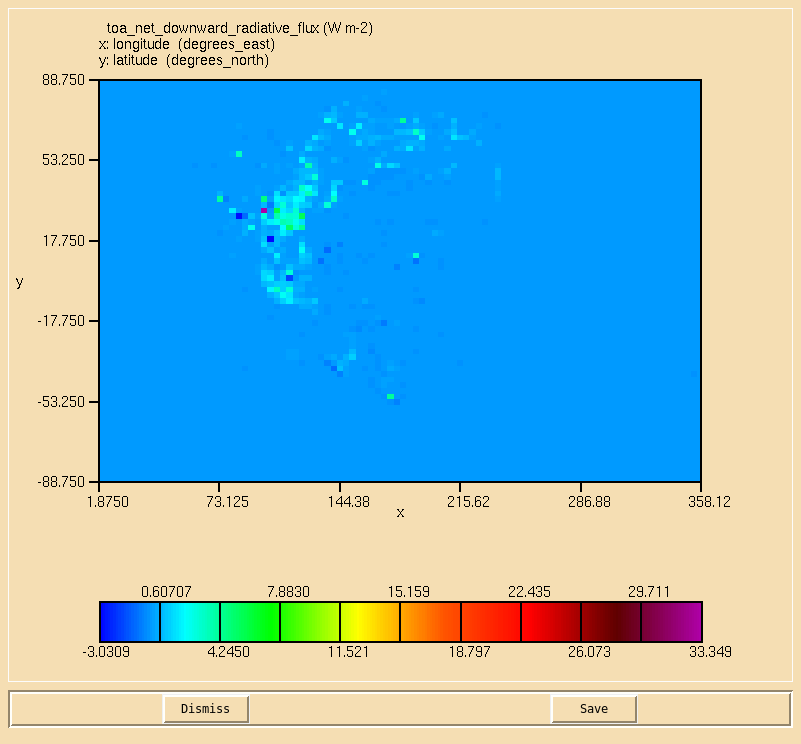
Image:Vn10.9_104_TOA.png|Net downward TOA radiative flux. |
|||
</gallery> |
|||
You were asked to |
|||
* ''Calculate the net downward top of the atmosphere radiative flux on the second radiation timestep.'' |
|||
You should use the python script provided to do this. |
|||
Sample output from this task can be found in the '''<code>/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.4/</code>''' directory on ARCHER, containing the following: |
Sample output from this task can be found in the '''<code>/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.4/</code>''' directory on ARCHER, containing the following: |
||
| Line 843: | Line 1,036: | ||
==Task 10.5: Calculate aerosol optical depth from the 3D aerosol extinction== |
==Task 10.5: Calculate aerosol optical depth from the 3D aerosol extinction== |
||
<span style="color:green">'''TASK 10.5:''' </span> |
<span style="color:green">'''TASK 10.5:''' Using the 3D aerosol extinction, calculate the 0.55 micron aerosol optical depth on the second radiation timestep, and compare this to your AOD from Task 10.2.</span> |
||
{| class="collapsible collapsed wikitable" |
|||
|- |
|||
! Hint |
|||
|- |
|||
| You will need to include contributions from both UKCA and CLASSIC (due to the dust scheme). |
|||
|- |
|||
| Remember to correctly calculate the grid-cell heights. |
|||
|} |
|||
===Python script=== |
|||
To calculate the AOD from the aerosol extinction, you will need to integrate this in the column. To do this you should first multiply by the height of each grid-cell before summing-up. |
|||
On ARCHER, an example python script to do this has been provided at '''<code>/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.5/calc_AOD.py</code>''': |
|||
#!/usr/bin/env python |
|||
# This file is part of the UKCA Tutorials: |
|||
# http://www.ukca.ac.uk/wiki/index.php/UKCA_Chemistry_and_Aerosol_Tutorials_at_vn10.9 |
|||
# Copyright (C) 2017 University of Cambridge |
|||
# This is free software: you can redistribute it and/or modify it under the |
|||
# terms of the GNU Lesser General Public License as published by the Free |
|||
# Software Foundation, either version 3 of the License, or (at your option) |
|||
# any later version. |
|||
# It is distributed in the hope that it will be useful, but WITHOUT ANY |
|||
# WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR |
|||
# A PARTICULAR PURPOSE. See the GNU Lesser General Public License for more |
|||
# details. |
|||
# You find a copy of the GNU Lesser General Public License at <http://www.gnu.org/licenses/>. |
|||
# Written by N. Luke Abraham 2017-12-11 <nla27@cam.ac.uk> |
|||
# preamble |
|||
import iris |
|||
import iris.time |
|||
import iris.analysis |
|||
import numpy as np |
|||
fname='/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.1/atmosa.pa19810901_00' |
|||
# constraint on time to get 2nd radiation timestep |
|||
tconstr=iris.Constraint(time=lambda cell: cell.point.hour == 2) |
|||
# load orography to enable correct calculation of level heights |
|||
orog=iris.load_cube( |
|||
'/work/n02/n02/hum/ancil/atmos/n48e/orography/globe30/v1/qrparm.orog', |
|||
iris.AttributeConstraint(STASH='m01s00i033')) |
|||
# load all extinction components at 0.55 micron |
|||
# must use this way of loading to account for constraint on time |
|||
with iris.FUTURE.context(cell_datetime_objects=True): |
|||
ukca=iris.load_cube(fname,[ |
|||
iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i530') & tconstr]) |
|||
classic=iris.load_cube(fname,[ |
|||
iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i540') & tconstr]) |
|||
# Calculate the correct height of each cell |
|||
# add the orography as an auxillary coordinate |
|||
auxcoord=iris.coords.AuxCoord(orog.data,standard_name=str(orog.standard_name),long_name="orography",var_name="orog",units=orog.units) |
|||
# added in to lat/lon (ht=0,lat=1,lon=2) |
|||
ukca.add_aux_coord(auxcoord,(1,2,)) |
|||
# now calculate the correct altitude above sea-level |
|||
factory=iris.aux_factory.HybridHeightFactory(delta=ukca.coord("level_height"),sigma=ukca.coord("sigma"),orography=ukca.coord("surface_altitude")) |
|||
# now create the 'altitude' derrived coordinate |
|||
ukca.add_aux_factory(factory) |
|||
# now calculate the height from the bounds |
|||
bounds = ukca.coord('altitude').bounds[:,:,:,1] - ukca.coord('altitude').bounds[:,:,:,0] |
|||
# mutliply by the height of each cell |
|||
ukca.data = ukca.data * bounds |
|||
classic.data = classic.data * bounds |
|||
# now sum up the column |
|||
ukca_int=ukca.collapsed('model_level_number',iris.analysis.SUM) |
|||
classic_int=classic.collapsed('model_level_number',iris.analysis.SUM) |
|||
# add together |
|||
aod=ukca_int.copy() |
|||
aod.data = ukca_int.data + classic_int.data |
|||
# rename |
|||
aod.rename('atmosphere_optical_thickness_due_to_aerosol') |
|||
# remove unlimited dimension when writing to netCDF |
|||
iris.FUTURE.netcdf_no_unlimited=True |
|||
# output to netCDF |
|||
iris.save(aod,'Task105_AOD.nc',netcdf_format='NETCDF3_CLASSIC') |
|||
On ARCHER, an example python script to calculate the difference from this AOD and the AOD calculated in Task10.2 is also provided at '''<code>/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.5/diff_AOD_methods.py</code>''': |
|||
#!/usr/bin/env python |
|||
# This file is part of the UKCA Tutorials: |
|||
# http://www.ukca.ac.uk/wiki/index.php/UKCA_Chemistry_and_Aerosol_Tutorials_at_vn10.9 |
|||
# Copyright (C) 2017 University of Cambridge |
|||
# This is free software: you can redistribute it and/or modify it under the |
|||
# terms of the GNU Lesser General Public License as published by the Free |
|||
# Software Foundation, either version 3 of the License, or (at your option) |
|||
# any later version. |
|||
# It is distributed in the hope that it will be useful, but WITHOUT ANY |
|||
# WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR |
|||
# A PARTICULAR PURPOSE. See the GNU Lesser General Public License for more |
|||
# details. |
|||
# You find a copy of the GNU Lesser General Public License at <http://www.gnu.org/licenses/>. |
|||
# Written by N. Luke Abraham 2017-12-11 <nla27@cam.ac.uk> |
|||
# preamble |
|||
import iris |
|||
dname='./Task102_AOD.nc' |
|||
cname='./Task105_AOD.nc' |
|||
diag=iris.load_cube(dname) |
|||
calc=iris.load_cube(cname) |
|||
# difference the fields |
|||
calc.data=calc.data - diag.data |
|||
# remove unlimited dimension when writing to netCDF |
|||
iris.FUTURE.netcdf_no_unlimited=True |
|||
# output to netCDF |
|||
iris.save(calc,'Task105_AOD_diff.nc',netcdf_format='NETCDF3_CLASSIC') |
|||
==Solution to Task 10.5 == |
==Solution to Task 10.5 == |
||
<gallery widths=300px heights=300px> |
|||
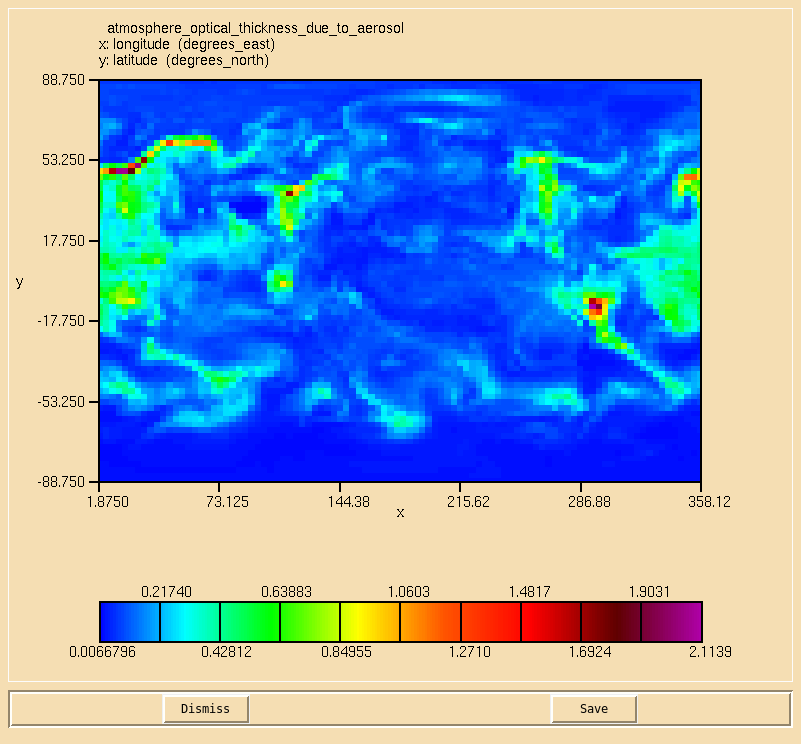
Image:Vn10.9_105_AOD.png|0.55 micron AOD calculated from the 3D extinction diagnostics. |
|||
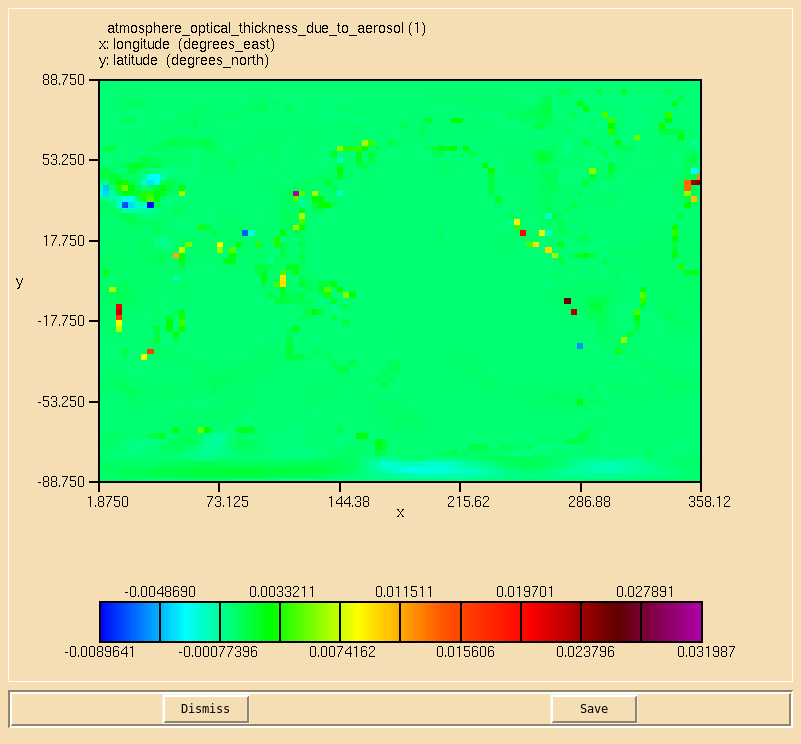
Image:Vn10.9_105_AOD_diff.png|Difference in AODs calculated by the two methods. |
|||
</gallery> |
|||
You were asked to |
|||
* ''Using the 3D aerosol extinction, calculate the 0.55 micron aerosol optical depth on the second radiation timestep, and compare this to your AOD from Task 10.2.'' |
|||
and were given the hints |
|||
:* ''You will need to include contributions from both UKCA and CLASSIC (due to the dust scheme).'' |
|||
:* ''Remember to correctly calculate the grid-cell heights.'' |
|||
You should use the python script provided to do this. |
|||
'''Note''' that there will be some small differences between these two fields, especially around coastlines. |
|||
Sample output from this task can be found in the '''<code>/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.5/</code>''' directory on ARCHER, containing the following: |
Sample output from this task can be found in the '''<code>/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.5/</code>''' directory on ARCHER, containing the following: |
||
| Line 855: | Line 1,199: | ||
==Task 10.6: Calculate the difference in aerosol impacts when <code>Sec_Org</code> is no longer formed from ALICE== |
==Task 10.6: Calculate the difference in aerosol impacts when <code>Sec_Org</code> is no longer formed from ALICE== |
||
<span style="color:green">'''TASK 10.6:''' </span> |
<span style="color:green">'''TASK 10.6:''' You should now remove the formation of '''<code>Sec_Org</code>''' from the chemical reaction added in Task 6.1, giving</span> |
||
<math> |
|||
==Solution to Task 10.5 == |
|||
\textrm{ALICE} + \textrm{OH} \longrightarrow \textrm{BOB} |
|||
</math> |
|||
<span style="color:green">and assess the impact this has on the aerosol diagnostics calculated in the previous tasks, 10.2, 10.3, and 10.4.</span> |
|||
{| class="collapsible collapsed wikitable" |
|||
|- |
|||
! Hint |
|||
|- |
|||
| Remember to remove <code>Sec_Org</code> from the diagnostics as well as the reaction. |
|||
|} |
|||
'''Note:''' as this a very short run (that is still spinning-up) and you will only be considering a single timestep, the results will be incredibly noisy. |
|||
==Code changes required to modify a reaction== |
|||
Modifying a reaction is very similar to [[UKCA_Chemistry_and_Aerosol_vn10.9_Tutorial_6#Adding_new_Chemical_Reactions|adding a new one]], although simpler as you are only changing what is already there. The exact routines that will need to be altered will depend on the reaction, but likely include: |
|||
===<code>ukca_chem_master.F90</code>=== |
|||
You must edit the master list of reactions, given by '''<code>ratb_defs_master</code>''' for bimolecular reactions, '''<code>ratt_defs_master</code>''' for termolecular reactions, '''<code>rath_defs_master</code>''' for heterogeneous reactions, or '''<code>ratj_defs_master</code>''' for photolysis reactions. |
|||
===<code>asad_bimol.F90</code>, <code>asad_trimol.F90</code>, <code>ukca_hetero_mod.F90</code>, <code>asad_hetero.F90</code>, <code>ukca_phot2d.F90</code>, and <code>ukca_strat_update.F90</code>=== |
|||
If you added any special code for the reaction(s) to <code>asad_bimol.F90</code>, <code>asad_trimol.F90</code>, <code>ukca_hetero_mod.F90</code>, <code>asad_hetero.F90</code>, <code>ukca_phot2d.F90</code>, or <code>ukca_strat_update.F90</code> it will need to be altered accordingly. The location of the reaction in the complete list is usually found by searching on the reactants and products, and if these change then the lookup will fail unless it is altered here too. |
|||
===<code>asad_flux_dat.F90</code>=== |
|||
If there are any diagnostics to output the reactions you are modifying, you may need to alter this specification as well if you have changed the reactants and/or products. |
|||
==Solution to Task 10.6 == |
|||
<gallery widths=300px heights=300px> |
|||
Image:Vn10.9_106_AOD_diff.png|Difference in AOD. |
|||
Image:Vn10.9_106_SSA_diff.png|Difference in single-scattering albedo. |
|||
Image:Vn10.9_106_TOA_diff.png|Difference in net downward TOA fluxes. |
|||
</gallery> |
|||
You were asked to |
|||
* ''You should now remove the formation of '''<code>Sec_Org</code>''' from the chemical reaction added in Task 6.1, giving'' |
|||
<math> |
|||
\textrm{ALICE} + \textrm{OH} \longrightarrow \textrm{BOB} |
|||
</math> |
|||
: ''and assess the impact this has on the aerosol diagnostics calculated in the previous tasks, 10.2, 10.3, and 10.4.'' |
|||
and were given the hint |
|||
:* ''Remember to remove <code>Sec_Org</code> from the diagnostics as well as the reaction.'' |
|||
You should use the above python scripts provided to do this. |
|||
For a working Rose suite that has completed this task, please see |
For a working Rose suite that has completed this task, please see |
||
| Line 943: | Line 1,340: | ||
==Checklist== |
==Checklist== |
||
: <span style="font-size:20px">☐</span> |
: <span style="font-size:20px">☐</span> Make new time and domain profiles as needed. |
||
: <span style="font-size:20px">☐</span> Run the TidyStashTransform transform macro. |
|||
: <span style="font-size:20px">☐</span> Output your diagnostics in STASH at: um <math>\rightarrow</math> namelist <math>\rightarrow</math> Model Input and Output <math>\rightarrow</math> STASH Requests and Profiles <math>\rightarrow</math> STASH Requests. |
|||
: <span style="font-size:20px">☐</span> When adding new aerosol diagnostics, remember to output on pseudo levels if appropriate. |
|||
: <span style="font-size:20px">☐</span> Run the TidyStashTransform transform macro again for the diagnostic requests. |
|||
: <span style="font-size:20px">☐</span> Save your suite. |
|||
: <span style="font-size:20px">☐</span> In the <code>roses/[SUITE-ID]</code> directory, run <code>fcm commit</code> to commit your changes to the repository. |
|||
: <span style="font-size:20px">☐</span> When modifying chemical reactions, remember to make changes for diagnostics and special cases, as well as altering <code>ukca_chem_master.F90</code>. |
|||
: <span style="font-size:20px">☐</span> The Iris python library can be used to process and visualise Unified Model output. |
|||
[[UKCA Chemistry and Aerosol vn10.9 Tutorial 11|Tutorial 11]] |
[[UKCA Chemistry and Aerosol vn10.9 Tutorial 11|Tutorial 11]] |
||
---- |
---- |
||
''Written by [[User:Nla27 | Luke Abraham]], Nicolas Bellouin, & Anja Schmidt 2017'' |
''Written by [[User:Nla27 | Luke Abraham]], Nicolas Bellouin, & Anja Schmidt 2017. With thanks to Ben Johnson.'' |
||
Latest revision as of 11:24, 7 May 2019
UKCA Chemistry and Aerosol Tutorials at vn10.9
What you will learn in this Tutorial
In this tutorial you will learn how to output and process aerosol diagnostics from UKCA.
Python
In this Tutorial you will make extensive use of Python to process the UM output. Instructions as to how to use this are in Tutorial 5.
Example python scripts are provided for each Task, and you should take the time to read through these and understand what they are doing. None of these scripts plot any of the output, so as an extension to these tasks you could try extending the scripts to plot the output from python directly, rather than visualising via Xconv. Further information (and examples) on how Iris plots can be found here:
- http://scitools.org.uk/iris/docs/latest/userguide/plotting_a_cube.html
- http://scitools.org.uk/iris/docs/latest/examples/General/global_map.html
Task 10.1: Output aerosol diagnostics
TASK 10.1: Output the following aerosol and radiation diagnostics to the UPA output stream. You should make a new time profile (called TRAD) to only output these on radiation timesteps. You will also need to make a new domain profile (called D3DAR) for s02i530 and s02i540, to output these on both model levels and pseudo levels.
| Hint |
|---|
| Remember to output your aerosol diagnostics on the AOT pseudo levels using the DIAGAOT profile. |
| STASH Section | STASH Item | STASH Name |
|---|---|---|
| 1 | 207 | INCOMING SW RAD FLUX (TOA): ALL TSS |
| 1 | 208 | OUTGOING SW RAD FLUX (TOA) |
| 2 | 205 | OUTGOING LW RAD FLUX (TOA) |
| 2 | 285 | MINERAL DUST OPTICAL DEPTH IN RADN. |
| 2 | 300 | AITKEN MODE (SOLUBLE) OPTICAL DEPTH |
| 2 | 301 | ACCUM MODE (SOLUBLE) OPTICAL DEPTH |
| 2 | 302 | COARSE MODE (SOLUBLE) OPTICAL DEPTH |
| 2 | 303 | AITKEN MODE (INSOL) OPTICAL DEPTH |
| 2 | 304 | ACCUM MODE (INSOL) OPTICAL DEPTH |
| 2 | 305 | COARSE MODE (INSOL) OPTICAL DEPTH |
| 2 | 585 | MINERAL DUST ABS. OPICAL DEPTH |
| 2 | 240 | AITKEN (SOLUBLE) ABS OPTICAL DEPTH |
| 2 | 241 | ACCUM (SOLUBLE) ABS OPTICAL DEPTH |
| 2 | 242 | COARSE (SOLUBLE) ABS OPTICAL DEPTH |
| 2 | 243 | AITKEN (INSOL) ABS OPTICAL DEPTH |
| 2 | 244 | ACCUM (INSOL) ABS OPTICAL DEPTH |
| 2 | 245 | COARSE (INSOL) ABS OPTICAL DEPTH |
| 2 | 530 | UKCA 3D AEROSOL EXTINCTION |
| 2 | 540 | CLASSIC 3D AEROSOL EXTINCTION |
Radiation Timesteps
The number of shortwave radiation timesteps per day is given by i_sw_radstep_perday_prog, found in um namelist UM Science Settings Section 01 - 02 Radiation Shortwave. The equivalent for longwave is i_lw_radstep_perday_prog, found in um namelist UM Science Settings Section 01 - 02 Radiation Longwave.
In the UKCA training suite, both of these are set to 16, i.e. a call every 90 minutes. This value is very configuration dependent - sometimes radiation is called every hour (24), sometimes every 3 hours (8) etc. You should check the settings in your suite if you need to consider any of the radiation diagnostics.
Making new STASH profiles
While we have covered outputting and creating new diagnostics in Tutorial 3, Tutorial 4, Tutorial 5, and Tutorial 9, you were only making use of existing profiles. Here you will learn how to make new time, domain, and usage profiles.
Time profiles
To make a new time profile, go to: um namelist Model Input and Output STASH Requests and Profiles Time Profiles. This contains a list of existing time profiles, with names like t3hmn_039ecafe etc.
To make a new time profile, you should:
- Right-click anywhere on the list and click the blue plus symbol (+) named new section
- This will make a new blank line (usually labelled 1, the next 2 etc.) with a red X next to it.
- Right-click this new line and click View namelist.
- Fill-in the values as required for what you want to do.
- When you have finished, you will need to run the When you have finished, you will need to run the stashindices.TidyStashTransform macro by going to the Metadata um drop-down menu. This will rename the new profile into the correct format.
Example: Output on Radiation Timesteps
You will have discovered that there are 16 radiation timesteps per day, but to work out how to output diagnostics you need to know many model timesteps per day there are. This can be found at: um namelist Top Level Model Control Model Domain and Timestep. The number is given by the variable steps_per_periodim, which is defined as the number of steps per period given (in seconds) by secs_per_periodim. For one day, this value is 86400.
In the UKCA training suite, secs_per_periodim=86400, and steps_per_periodim=48. This means that there are 48 timesteps per day, i.e. each timestep is 30 minutes.
We can combine this with the number of radiation timesteps per day (16), to calculate that radiation is called every third model timestep (48/16 = 3) in this configuration. We can now fill-in the values in the new time profile accordingly, e.g.
ityp: No time processingtim_name: e.g. TRADunt3: Timesteps (1)iopt: Regular intervalsistr: 1 (i.e. start on first timestep)iend: -1 (i.e. never stop outputting)ifre: 3 (i.e. output every third timestep)
We need to start at timestep 1, as radiation is called on the first timestep, and then called every third model timestep after that. In contrast, the UKCA Newton-Raphson chemical solver is first called on the first hour, and then every hour after that. For the UKCA training suite this would mean that istr=2 and ifre=2.
Domain profiles
To make a new time profile, go to: um namelist Model Input and Output STASH Requests and Profiles Domain Profiles. This contains a list of existing domain profiles, with names like dallth_1e2e730d etc.
To make a new domain profile, you should:
- Right-click anywhere on the list and click the blue plus symbol (+) named new section
- This will make a new blank line (usually labelled 1, the next 2 etc.) with a red X next to it.
- Right-click this new line and click View namelist.
- Fill-in the values as required for what you want to do.
- When you have finished, you will need to run the When you have finished, you will need to run the stashindices.TidyStashTransform macro by going to the Metadata um drop-down menu. This will rename the new profile into the correct format.
Example: Output AOT pseudo levels on all model theta levels
Aerosol diagnostics are defined on a series of 6 pseudo levels, corresponding to 0.38, 0.44, 0.55, 0.67, 0.87, and 1.02 microns. You will therefore need to output on your data on all of these. For some diagnostics, such as AOD, these are defined on a single level, but others, such as extinction, are a 3D field over the whole atmosphere. Therefore, to output the 3D aerosol extinction we need to make a new profile over all model (theta grid) levels, and all 6 pseudo levels.
We can now fill-in the values in the new domain profile accordingly, e.g.
dom_name: e.g. D3DARiopl: Variable derived on model theta levels (Charney-Phillips Grid) (2)ilevs: provide range [bottom and top level numbers]ilevb: 1 (i.e. 20m level)ilevt: 85 (i.e. highest level)plt: Radiation bands for calculating aerosol optical depth (4)
- This will then insert a new option below (
pslist) to allow you to specify the bands you require.
pslist: press the blue plus symbol (+) 5 times, and enter the numbers 1 to 6 in each of the boxes to output all AOT pseudo levels.iopa: Full model area (1)imsk: Land and sea pointsimn: None (0)iwt: Nonets: false
Usage profiles
Making new usage profiles is not as straight-forward as for time and domain profiles, as these are used to output diagnostics to different output files (and possibly to climate mean files). To see what output streams are available, go to: um namelist Model Input and Output Model Output Streams. In the UKCA training suite, only pp0 is active.
To look at climate meaning, go to: um namelist Model Input and Output Dumping and Meaning. This is controlled by the logical l_meaning_sequence, which in the UKCA training suite is set to false (i.e. no climate meaning). If this is set to true then various settings will need to be chosen, such as the reference time, the frequency of files, and which files to output.
To view the usage profiles, go to: um namelist Model Input and Output STASH Requests and Profiles Usage Profiles. This contains a list of existing domain profiles, with names like upa_ffb3f00b etc. If you look at the upa namelist (by right-clicking on it and clicking view namelist) you can see that this corresponds with the pp0 output stream.
It is also possible to write diagnostics to a tag, which can then be read by other parts of the code. This is how UKCA couples to the UM, via tag=98. This can be seen in the upukca usage profile.
When you take a copy of an existing configuration it is unlikely that you will need to make any changes to any (or make new) usage profiles. However, if you do want to make a new usage profile, you should:
- Right-click anywhere on the list and click the blue plus symbol (+) named new section
- This will make a new blank line (usually labelled 1, the next 2 etc.) with a red X next to it.
- Right-click this new line and click View namelist.
- Fill-in the values as required for what you want to do.
- When you have finished, you will need to run the When you have finished, you will need to run the stashindices.TidyStashTransform macro by going to the Metadata um drop-down menu. This will rename the new profile into the correct format.
Note that if you are making a new usage profile that is not associated with a tag, you may also need to make changes to the output streams. This could be quite involved.
Using your new profiles
Once you have made your new profiles you should then use them in the usual way when outputting diagnostics, as covered in Tutorial 3.
When all your new STASH is set-up, you can use the Validator Macros to check if the names are correct, or if there are any unused STASH items etc. To run this, go to Metadata Check all Validator Macros from the drop-down menu. You can also check for fail-if or warn-if issues. However, these won't warn of incorrect domain requests (i.e. requesting pressure-levels for diagnostics only defined on a single level), so some care should still be taken.
Solution to Task 10.1
You were given the task
- Output the following aerosol and radiation diagnostics to the UPA output stream. You should make a new time profile (called TRAD) to only output these on radiation timesteps. You will also need to make a new domain profile (called D3DAR) for s02i530 and s02i540, to output these on both model levels and pseudo levels.
| STASH Section | STASH Item | STASH Name |
|---|---|---|
| 1 | 207 | INCOMING SW RAD FLUX (TOA): ALL TSS |
| 1 | 208 | OUTGOING SW RAD FLUX (TOA) |
| 2 | 205 | OUTGOING LW RAD FLUX (TOA) |
| 2 | 285 | MINERAL DUST OPTICAL DEPTH IN RADN. |
| 2 | 300 | AITKEN MODE (SOLUBLE) OPTICAL DEPTH |
| 2 | 301 | ACCUM MODE (SOLUBLE) OPTICAL DEPTH |
| 2 | 302 | COARSE MODE (SOLUBLE) OPTICAL DEPTH |
| 2 | 303 | AITKEN MODE (INSOL) OPTICAL DEPTH |
| 2 | 304 | ACCUM MODE (INSOL) OPTICAL DEPTH |
| 2 | 305 | COARSE MODE (INSOL) OPTICAL DEPTH |
| 2 | 585 | MINERAL DUST ABS. OPICAL DEPTH |
| 2 | 240 | AITKEN (SOLUBLE) ABS OPTICAL DEPTH |
| 2 | 241 | ACCUM (SOLUBLE) ABS OPTICAL DEPTH |
| 2 | 242 | COARSE (SOLUBLE) ABS OPTICAL DEPTH |
| 2 | 243 | AITKEN (INSOL) ABS OPTICAL DEPTH |
| 2 | 244 | ACCUM (INSOL) ABS OPTICAL DEPTH |
| 2 | 245 | COARSE (INSOL) ABS OPTICAL DEPTH |
| 2 | 530 | UKCA 3D AEROSOL EXTINCTION |
| 2 | 540 | CLASSIC 3D AEROSOL EXTINCTION |
and were given the hint
- Remember to output your aerosol diagnostics on the AOT pseudo levels using the DIAGAOT profile.
For a working Rose suite that has completed this task, please see
- ARCHER:
u-as292@62651 - vm:
u-as297@62631
The specific Rose changes made are:
- ARCHER: https://code.metoffice.gov.uk/trac/roses-u/changeset/62651/a/s/2/9/2/trunk
- vm: https://code.metoffice.gov.uk/trac/roses-u/changeset/62631/a/s/2/9/7/trunk
The specific Rose changes made are:
ARCHER:
Index: app/um/rose-app.conf =================================================================== --- app/um/rose-app.conf (revision 60289) +++ app/um/rose-app.conf (revision 62651) @@ -2744,6 +2744,40 @@ precip_segment_size=32 ukca_mode_seg_size=4 +[namelist:umstash_domain(d3dar_72578706)] +dom_name='D3DAR' +!!iest=0 +ilevs=1 +imn=0 +imsk=1 +!!inth=0 +iopa=1 +iopl=2 +!!isth=0 +!!iwst=0 +iwt=0 +!!l_spml_ts=.false. +levb=01 +!!levlst=0 +levt=85 +plt=4 +pslist=1,2,3,4,5,6 +!!rlevlst=0 +!!spml_bot=0 +!!spml_ew=0 +!!spml_ns=0 +!!spml_top=0 +!!tblim=0 +!!tblimr=0 +!!telim=0 +!!tnlim=0 +ts=.false. +!!tslim=0 +!!tsnum=0 +!!ttlim=0 +!!ttlimr=0 +!!twlim=0 + [namelist:umstash_domain(dallrh_0496a967)] dom_name='DALLRH' !!iest=0 @@ -3025,6 +3059,22 @@ tim_name='TALLTS' use_name='UPUKCA' +[namelist:umstash_streq(01207_ed72c304)] +dom_name='DIAG' +isec=1 +item=207 +package= +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(01208_83140cd8)] +dom_name='DIAG' +isec=1 +item=208 +package= +tim_name='TRAD' +use_name='UPA' + [namelist:umstash_streq(01235_3511dd9f)] dom_name='DIAG' isec=1 @@ -3033,14 +3083,142 @@ tim_name='TALLTS' use_name='UPUKCA' -[namelist:umstash_streq(02301_0f7c5f4a)] +[namelist:umstash_streq(02205_357bf644)] +dom_name='DIAG' +isec=2 +item=205 +package= +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02240_d97aaab7)] dom_name='DIAGAOT' isec=2 +item=240 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02241_8cda3169)] +dom_name='DIAGAOT' +isec=2 +item=241 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02242_91e371db)] +dom_name='DIAGAOT' +isec=2 +item=242 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02243_21bc5f11)] +dom_name='DIAGAOT' +isec=2 +item=243 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02244_32fe0790)] +dom_name='DIAGAOT' +isec=2 +item=244 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02245_da00b6ef)] +dom_name='DIAGAOT' +isec=2 +item=245 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02285_1d9800f0)] +dom_name='DIAGAOT' +isec=2 +item=285 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02300_8b9907b5)] +dom_name='DIAGAOT' +isec=2 +item=300 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02301_b8711d23)] +dom_name='DIAGAOT' +isec=2 item=301 -package='UKCA Testing' -tim_name='T3HMN' +package=' ' +tim_name='TRAD' use_name='UPA' +[namelist:umstash_streq(02302_420d0ec7)] +dom_name='DIAGAOT' +isec=2 +item=302 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02303_ad5c3af4)] +dom_name='DIAGAOT' +isec=2 +item=303 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02304_8c1869b6)] +dom_name='DIAGAOT' +isec=2 +item=304 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02305_9ecd020a)] +dom_name='DIAGAOT' +isec=2 +item=305 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02530_7a218781)] +dom_name='D3DAR' +isec=2 +item=530 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02540_56484a28)] +dom_name='D3DAR' +isec=2 +item=540 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02585_38287367)] +dom_name='DIAGAOT' +isec=2 +item=585 +package=' ' +tim_name='TRAD' +use_name='UPA' + [namelist:umstash_streq(03025_c8768f77)] dom_name='DIAG' isec=3 @@ -3551,6 +3729,25 @@ !!unt2=2 unt3=1 +[namelist:umstash_time(trad_4c3a45a7)] +!!iedt=0 +iend=-1 +ifre=3 +!!intv=0 +!!ioff=0 +iopt=1 +!!isam=0 +!!isdt=0 +!!iser=0 +istr=1 +!!itimes=0 +ityp=1 +!!lts0=.false. +tim_name='TRAD' +!!unt1=2 +!!unt2=2 +unt3=1 + [namelist:umstash_use(upa_ffb3f00b)] file_id='pp0' locn=3
These differences can be found in the file /home/ukca/Tutorial/vn10.9/worked_solutions/Task10.1/Task10.1_rose.patch on PUMA.
vm:
Index: app/um/rose-app.conf =================================================================== --- app/um/rose-app.conf (revision 60286) +++ app/um/rose-app.conf (revision 62631) @@ -2849,6 +2849,40 @@ !!ttlimr=0.0 !!twlim=0 +[namelist:umstash_domain(d3dar_72578706)] +dom_name='D3DAR' +!!iest=0 +ilevs=1 +imn=0 +imsk=1 +!!inth=0 +iopa=1 +iopl=2 +!!isth=0 +!!iwst=0 +iwt=0 +!!l_spml_ts=.false. +levb=01 +!!levlst=0 +levt=85 +plt=4 +pslist=1,2,3,4,5,6 +!!rlevlst=0 +!!spml_bot=0 +!!spml_ew=0 +!!spml_ns=0 +!!spml_top=0 +!!tblim=0 +!!tblimr=0 +!!telim=0 +!!tnlim=0 +ts=.false. +!!tslim=0 +!!tsnum=0 +!!ttlim=0 +!!ttlimr=0 +!!twlim=0 + [namelist:umstash_domain(dallrh_0496a967)] dom_name='DALLRH' !!iest=0 @@ -3617,6 +3651,22 @@ tim_name='TALLTS' use_name='UPUKCA' +[namelist:umstash_streq(01207_ed72c304)] +dom_name='DIAG' +isec=1 +item=207 +package= +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(01208_83140cd8)] +dom_name='DIAG' +isec=1 +item=208 +package= +tim_name='TRAD' +use_name='UPA' + [namelist:umstash_streq(01235_3511dd9f)] dom_name='DIAG' isec=1 @@ -3625,14 +3675,142 @@ tim_name='TALLTS' use_name='UPUKCA' -[namelist:umstash_streq(02301_0f7c5f4a)] +[namelist:umstash_streq(02205_357bf644)] +dom_name='DIAG' +isec=2 +item=205 +package= +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02240_d97aaab7)] dom_name='DIAGAOT' isec=2 +item=240 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02241_8cda3169)] +dom_name='DIAGAOT' +isec=2 +item=241 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02242_91e371db)] +dom_name='DIAGAOT' +isec=2 +item=242 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02243_21bc5f11)] +dom_name='DIAGAOT' +isec=2 +item=243 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02244_32fe0790)] +dom_name='DIAGAOT' +isec=2 +item=244 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02245_da00b6ef)] +dom_name='DIAGAOT' +isec=2 +item=245 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02285_1d9800f0)] +dom_name='DIAGAOT' +isec=2 +item=285 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02300_8b9907b5)] +dom_name='DIAGAOT' +isec=2 +item=300 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02301_b8711d23)] +dom_name='DIAGAOT' +isec=2 item=301 -package='UKCA Testing' -tim_name='T3HMN' +package=' ' +tim_name='TRAD' use_name='UPA' +[namelist:umstash_streq(02302_420d0ec7)] +dom_name='DIAGAOT' +isec=2 +item=302 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02303_ad5c3af4)] +dom_name='DIAGAOT' +isec=2 +item=303 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02304_8c1869b6)] +dom_name='DIAGAOT' +isec=2 +item=304 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02305_9ecd020a)] +dom_name='DIAGAOT' +isec=2 +item=305 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02530_7a218781)] +dom_name='D3DAR' +isec=2 +item=530 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02540_56484a28)] +dom_name='D3DAR' +isec=2 +item=540 +package=' ' +tim_name='TRAD' +use_name='UPA' + +[namelist:umstash_streq(02585_38287367)] +dom_name='DIAGAOT' +isec=2 +item=585 +package=' ' +tim_name='TRAD' +use_name='UPA' + [namelist:umstash_streq(03025_c8768f77)] dom_name='DIAG' isec=3 @@ -4504,6 +4682,25 @@ unt2=2 unt3=4 +[namelist:umstash_time(trad_4c3a45a7)] +!!iedt=0 +iend=-1 +ifre=3 +!!intv=0 +!!ioff=0 +iopt=1 +!!isam=0 +!!isdt=0 +!!iser=0 +istr=1 +!!itimes=0 +ityp=1 +!!lts0=.false. +tim_name='TRAD' +!!unt1=2 +!!unt2=2 +unt3=1 + [namelist:umstash_time(traddm_fa7c24ce)] !!iedt=0 iend=-1
If you open the .pa file in Xconv, you should see the following additional fields:
0 : 96 72 1 2 field200: INCOMING SW RAD FLUX (TOA): ALL TSS 1 : 96 72 1 2 field201: OUTGOING SW RAD FLUX (TOA) 2 : 96 72 1 2 olr: OUTGOING LW RAD FLUX (TOA) 3 : 96 72 6 2 unspecified: Stash code = 2240 4 : 96 72 6 2 unspecified: Stash code = 2241 5 : 96 72 6 2 unspecified: Stash code = 2242 6 : 96 72 6 2 unspecified: Stash code = 2243 7 : 96 72 6 2 unspecified: Stash code = 2244 8 : 96 72 6 2 unspecified: Stash code = 2245 9 : 96 72 6 2 unspecified: MINERAL DUST OPTICAL DEPTH IN RADN. 10 : 96 72 6 2 unspecified: AITKEN MODE (SOLUBLE) OPTICAL DEPTH 11 : 96 72 6 2 unspecified: ACCUM MODE (SOLUBLE) OPTICAL DEPTH 12 : 96 72 6 2 unspecified: COARSE MODE (SOLUBLE) OPTICAL DEPTH 13 : 96 72 6 2 unspecified: AITKEN MODE (INSOL) OPTICAL DEPTH 14 : 96 72 6 2 unspecified: ACCUM MODE (INSOL) OPTICAL DEPTH 15 : 96 72 6 2 unspecified: COARSE MODE (INSOL) OPTICAL DEPTH 16 : 96 72 38 2 unspecified: Stash code = 2530 17 : 96 72 38 2 unspecified: Stash code = 2530 18 : 96 72 38 2 unspecified: Stash code = 2530 19 : 96 72 38 2 unspecified: Stash code = 2530 20 : 96 72 38 2 unspecified: Stash code = 2530 21 : 96 72 38 2 unspecified: Stash code = 2530 22 : 96 72 38 2 unspecified: Stash code = 2540 23 : 96 72 38 2 unspecified: Stash code = 2540 24 : 96 72 38 2 unspecified: Stash code = 2540 25 : 96 72 38 2 unspecified: Stash code = 2540 26 : 96 72 38 2 unspecified: Stash code = 2540 27 : 96 72 38 2 unspecified: Stash code = 2540 28 : 96 72 6 2 unspecified: Stash code = 2585
Sample output from this task can be found at /work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.1/atmosa.pa19810901_00 on ARCHER.
Task 10.2: Calculate aerosol optical depth
TASK 10.2: Calculate the aerosol optical depth at 0.55 microns on the second radiation timestep.
| Hint |
|---|
| The UM calculates optical depth diagnostics on 6 pseudo levels corresponding to 0.38, 0.44, 0.55, 0.67, 0.87, and 1.02 microns, therefore 0.55 microns is pseudo level 3. |
Note: You will need to use the CLASSIC mineral dust optical depth diagnostic, as the configuration used in these tutorials does not use modal dust. Likewise, the insoluble accumulation and coarse mode diagnostics will be zero as these modes are not used in the configuration used here.
Python script
To calculate the total aerosol optical depth at 0.55 microns, you should sum up the contribution from the different aerosol components.
On ARCHER, an example python script to do this has been provided at /work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.2/write_AOD.py:
#!/usr/bin/env python # This file is part of the UKCA Tutorials: # http://www.ukca.ac.uk/wiki/index.php/UKCA_Chemistry_and_Aerosol_Tutorials_at_vn10.9 # Copyright (C) 2017 University of Cambridge # This is free software: you can redistribute it and/or modify it under the # terms of the GNU Lesser General Public License as published by the Free # Software Foundation, either version 3 of the License, or (at your option) # any later version. # It is distributed in the hope that it will be useful, but WITHOUT ANY # WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR # A PARTICULAR PURPOSE. See the GNU Lesser General Public License for more # details. # You find a copy of the GNU Lesser General Public License at <http://www.gnu.org/licenses/>. # Written by N. Luke Abraham 2017-12-11 <nla27@cam.ac.uk> # preamble import iris import iris.time fname='/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.1/atmosa.pa19810901_00' # constraint on time to get 2nd radiation timestep tconstr=iris.Constraint(time=lambda cell: cell.point.hour == 2) # load all AOD components at 0.55 micron # must use this way of loading to account for constraint on time with iris.FUTURE.context(cell_datetime_objects=True): aod=iris.load(fname,[ iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i285') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i300') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i301') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i302') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i303') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i304') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i305') & tconstr]) # make cube to store total AOD aodsum=aod[0].copy() # add-up components aodsum.data=aod[0].data+aod[1].data+aod[2].data+aod[3].data+aod[4].data+aod[5].data+aod[6].data # rename aodsum.rename('atmosphere_optical_thickness_due_to_aerosol') # remove unlimited dimension when writing to netCDF iris.FUTURE.netcdf_no_unlimited=True # output to netCDF iris.save(aodsum,'Task102_AOD.nc',netcdf_format='NETCDF3_CLASSIC')
Solution to Task 10.2
You were asked to
- Calculate the aerosol optical depth at 0.55 microns on the second radiation timestep.
and were given the hint
- The UM calculates optical depth diagnostics on 6 pseudo levels corresponding to 0.38, 0.44, 0.55, 0.67, 0.87, and 1.02 microns, therefore 0.55 microns is pseudo level 3.
You should make use of the python script provided to do this.
Sample output from this task can be found in the /work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.2/ directory on ARCHER, containing the following:
Task102_AOD.nc write_AOD.py
Task 10.3: Calculate the single-scattering albedo
TASK 10.3: Calculate the single-scattering albedo at 0.55 microns on the second radiation timestep, defined as:
Python script
To calculate the single-scattering albedo at 0.55 microns, you should sum up the contribution to AAOD and AOD from the different aerosol components.
On ARCHER, an example python script to do this has been provided at /work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.3/write_SSA.py:
#!/usr/bin/env python # This file is part of the UKCA Tutorials: # http://www.ukca.ac.uk/wiki/index.php/UKCA_Chemistry_and_Aerosol_Tutorials_at_vn10.9 # Copyright (C) 2017 University of Cambridge # This is free software: you can redistribute it and/or modify it under the # terms of the GNU Lesser General Public License as published by the Free # Software Foundation, either version 3 of the License, or (at your option) # any later version. # It is distributed in the hope that it will be useful, but WITHOUT ANY # WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR # A PARTICULAR PURPOSE. See the GNU Lesser General Public License for more # details. # You find a copy of the GNU Lesser General Public License at <http://www.gnu.org/licenses/>. # Written by N. Luke Abraham 2017-12-11 <nla27@cam.ac.uk> # preamble import iris import iris.time fname='/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.1/atmosa.pa19810901_00' # constraint on time to get 2nd radiation timestep tconstr=iris.Constraint(time=lambda cell: cell.point.hour == 2) # load all AOD & AAOD components at 0.55 micron # must use this way of loading to account for constraint on time with iris.FUTURE.context(cell_datetime_objects=True): aod=iris.load(fname,[ iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i285') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i300') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i301') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i302') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i303') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i304') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i305') & tconstr]) aaod=iris.load(fname,[ iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i585') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i240') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i241') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i242') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i243') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i244') & tconstr, iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i245') & tconstr]) # make cube to store total AOD aodsum=aod[0].copy() # add-up components aodsum.data=aod[0].data+aod[1].data+aod[2].data+aod[3].data+aod[4].data+aod[5].data+aod[6].data # make cube to store total AAOD aaodsum=aaod[0].copy() # add-up components aaodsum.data=aaod[0].data+aaod[1].data+aaod[2].data+aaod[3].data+aaod[4].data+aaod[5].data+aaod[6].data # calculate single-scattering albedo ssa=aodsum.copy() ssa.data = 1.0 - (aaodsum.data/aodsum.data) # rename ssa.rename('single_scattering_albedo_in_air_due_to_ambient_aerosol_particles') # remove unlimited dimension when writing to netCDF iris.FUTURE.netcdf_no_unlimited=True # output to netCDF iris.save(ssa,'Task103_SSA.nc',netcdf_format='NETCDF3_CLASSIC')
Solution to Task 10.3
You were asked to
- Calculate the single-scattering albedo at 0.55 microns on the second radiation timestep, defined as:
You should use the python script provided to do this.
Sample output from this task can be found in the /work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.3/ directory on ARCHER, containing the following:
Task103_SSA.nc write_SSA.py
Task 10.4: Calculate the top of the atmosphere net downward radiative flux
TASK 10.4: Calculate the net downward top of the atmosphere radiative flux on the second radiation timestep.
Python script
To calculate the net TOA downward radiative flux, you should sum up the outgoing contributions from shortwave and longwave radiation, and take this away from the incoming shortwave radiative flux.
On ARCHER, an example python script to do this has been provided at /work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.4/write_TOA.py:
#!/usr/bin/env python # This file is part of the UKCA Tutorials: # http://www.ukca.ac.uk/wiki/index.php/UKCA_Chemistry_and_Aerosol_Tutorials_at_vn10.9 # Copyright (C) 2017 University of Cambridge # This is free software: you can redistribute it and/or modify it under the # terms of the GNU Lesser General Public License as published by the Free # Software Foundation, either version 3 of the License, or (at your option) # any later version. # It is distributed in the hope that it will be useful, but WITHOUT ANY # WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR # A PARTICULAR PURPOSE. See the GNU Lesser General Public License for more # details. # You find a copy of the GNU Lesser General Public License at <http://www.gnu.org/licenses/>. # Written by N. Luke Abraham 2017-12-11 <nla27@cam.ac.uk> # preamble import iris import iris.time fname='/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.1/atmosa.pa19810901_00' # constraint on time to get 2nd radiation timestep tconstr=iris.Constraint(time=lambda cell: cell.point.hour == 2) # load all TOA components at 0.55 micron # must use this way of loading to account for constraint on time with iris.FUTURE.context(cell_datetime_objects=True): isw=iris.load_cube(fname,[iris.AttributeConstraint(STASH='m01s01i207') & tconstr]) osw=iris.load_cube(fname,[iris.AttributeConstraint(STASH='m01s01i208') & tconstr]) olw=iris.load_cube(fname,[iris.AttributeConstraint(STASH='m01s02i205') & tconstr]) # make cube to store net downward TOA flux toa=isw.copy() # add-up components toa.data=isw.data - (osw.data + olw.data) toa.rename('toa_net_downward_radiative_flux') # remove unlimited dimension when writing to netCDF iris.FUTURE.netcdf_no_unlimited=True # output to netCDF iris.save(toa,'Task104_TOA.nc',netcdf_format='NETCDF3_CLASSIC')
Solution to Task 10.4
You were asked to
- Calculate the net downward top of the atmosphere radiative flux on the second radiation timestep.
You should use the python script provided to do this.
Sample output from this task can be found in the /work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.4/ directory on ARCHER, containing the following:
Task104_TOA.nc write_TOA.py
Task 10.5: Calculate aerosol optical depth from the 3D aerosol extinction
TASK 10.5: Using the 3D aerosol extinction, calculate the 0.55 micron aerosol optical depth on the second radiation timestep, and compare this to your AOD from Task 10.2.
| Hint |
|---|
| You will need to include contributions from both UKCA and CLASSIC (due to the dust scheme). |
| Remember to correctly calculate the grid-cell heights. |
Python script
To calculate the AOD from the aerosol extinction, you will need to integrate this in the column. To do this you should first multiply by the height of each grid-cell before summing-up.
On ARCHER, an example python script to do this has been provided at /work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.5/calc_AOD.py:
#!/usr/bin/env python # This file is part of the UKCA Tutorials: # http://www.ukca.ac.uk/wiki/index.php/UKCA_Chemistry_and_Aerosol_Tutorials_at_vn10.9 # Copyright (C) 2017 University of Cambridge # This is free software: you can redistribute it and/or modify it under the # terms of the GNU Lesser General Public License as published by the Free # Software Foundation, either version 3 of the License, or (at your option) # any later version. # It is distributed in the hope that it will be useful, but WITHOUT ANY # WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR # A PARTICULAR PURPOSE. See the GNU Lesser General Public License for more # details. # You find a copy of the GNU Lesser General Public License at <http://www.gnu.org/licenses/>. # Written by N. Luke Abraham 2017-12-11 <nla27@cam.ac.uk> # preamble import iris import iris.time import iris.analysis import numpy as np fname='/work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.1/atmosa.pa19810901_00' # constraint on time to get 2nd radiation timestep tconstr=iris.Constraint(time=lambda cell: cell.point.hour == 2) # load orography to enable correct calculation of level heights orog=iris.load_cube( '/work/n02/n02/hum/ancil/atmos/n48e/orography/globe30/v1/qrparm.orog', iris.AttributeConstraint(STASH='m01s00i033')) # load all extinction components at 0.55 micron # must use this way of loading to account for constraint on time with iris.FUTURE.context(cell_datetime_objects=True): ukca=iris.load_cube(fname,[ iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i530') & tconstr]) classic=iris.load_cube(fname,[ iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i540') & tconstr]) # Calculate the correct height of each cell # add the orography as an auxillary coordinate auxcoord=iris.coords.AuxCoord(orog.data,standard_name=str(orog.standard_name),long_name="orography",var_name="orog",units=orog.units) # added in to lat/lon (ht=0,lat=1,lon=2) ukca.add_aux_coord(auxcoord,(1,2,)) # now calculate the correct altitude above sea-level factory=iris.aux_factory.HybridHeightFactory(delta=ukca.coord("level_height"),sigma=ukca.coord("sigma"),orography=ukca.coord("surface_altitude")) # now create the 'altitude' derrived coordinate ukca.add_aux_factory(factory) # now calculate the height from the bounds bounds = ukca.coord('altitude').bounds[:,:,:,1] - ukca.coord('altitude').bounds[:,:,:,0] # mutliply by the height of each cell ukca.data = ukca.data * bounds classic.data = classic.data * bounds # now sum up the column ukca_int=ukca.collapsed('model_level_number',iris.analysis.SUM) classic_int=classic.collapsed('model_level_number',iris.analysis.SUM) # add together aod=ukca_int.copy() aod.data = ukca_int.data + classic_int.data # rename aod.rename('atmosphere_optical_thickness_due_to_aerosol') # remove unlimited dimension when writing to netCDF iris.FUTURE.netcdf_no_unlimited=True # output to netCDF iris.save(aod,'Task105_AOD.nc',netcdf_format='NETCDF3_CLASSIC')
On ARCHER, an example python script to calculate the difference from this AOD and the AOD calculated in Task10.2 is also provided at /work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.5/diff_AOD_methods.py:
#!/usr/bin/env python # This file is part of the UKCA Tutorials: # http://www.ukca.ac.uk/wiki/index.php/UKCA_Chemistry_and_Aerosol_Tutorials_at_vn10.9 # Copyright (C) 2017 University of Cambridge # This is free software: you can redistribute it and/or modify it under the # terms of the GNU Lesser General Public License as published by the Free # Software Foundation, either version 3 of the License, or (at your option) # any later version. # It is distributed in the hope that it will be useful, but WITHOUT ANY # WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR # A PARTICULAR PURPOSE. See the GNU Lesser General Public License for more # details. # You find a copy of the GNU Lesser General Public License at <http://www.gnu.org/licenses/>. # Written by N. Luke Abraham 2017-12-11 <nla27@cam.ac.uk> # preamble import iris dname='./Task102_AOD.nc' cname='./Task105_AOD.nc' diag=iris.load_cube(dname) calc=iris.load_cube(cname) # difference the fields calc.data=calc.data - diag.data # remove unlimited dimension when writing to netCDF iris.FUTURE.netcdf_no_unlimited=True # output to netCDF iris.save(calc,'Task105_AOD_diff.nc',netcdf_format='NETCDF3_CLASSIC')
Solution to Task 10.5
You were asked to
- Using the 3D aerosol extinction, calculate the 0.55 micron aerosol optical depth on the second radiation timestep, and compare this to your AOD from Task 10.2.
and were given the hints
- You will need to include contributions from both UKCA and CLASSIC (due to the dust scheme).
- Remember to correctly calculate the grid-cell heights.
You should use the python script provided to do this.
Note that there will be some small differences between these two fields, especially around coastlines.
Sample output from this task can be found in the /work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.5/ directory on ARCHER, containing the following:
Task105_AOD.nc Task105_AOD_diff.nc calc_AOD.py diff_AOD_methods.py
Task 10.6: Calculate the difference in aerosol impacts when Sec_Org is no longer formed from ALICE
TASK 10.6: You should now remove the formation of Sec_Org from the chemical reaction added in Task 6.1, giving
and assess the impact this has on the aerosol diagnostics calculated in the previous tasks, 10.2, 10.3, and 10.4.
| Hint |
|---|
Remember to remove Sec_Org from the diagnostics as well as the reaction.
|
Note: as this a very short run (that is still spinning-up) and you will only be considering a single timestep, the results will be incredibly noisy.
Code changes required to modify a reaction
Modifying a reaction is very similar to adding a new one, although simpler as you are only changing what is already there. The exact routines that will need to be altered will depend on the reaction, but likely include:
ukca_chem_master.F90
You must edit the master list of reactions, given by ratb_defs_master for bimolecular reactions, ratt_defs_master for termolecular reactions, rath_defs_master for heterogeneous reactions, or ratj_defs_master for photolysis reactions.
asad_bimol.F90, asad_trimol.F90, ukca_hetero_mod.F90, asad_hetero.F90, ukca_phot2d.F90, and ukca_strat_update.F90
If you added any special code for the reaction(s) to asad_bimol.F90, asad_trimol.F90, ukca_hetero_mod.F90, asad_hetero.F90, ukca_phot2d.F90, or ukca_strat_update.F90 it will need to be altered accordingly. The location of the reaction in the complete list is usually found by searching on the reactants and products, and if these change then the lookup will fail unless it is altered here too.
asad_flux_dat.F90
If there are any diagnostics to output the reactions you are modifying, you may need to alter this specification as well if you have changed the reactants and/or products.
Solution to Task 10.6
You were asked to
- You should now remove the formation of
Sec_Orgfrom the chemical reaction added in Task 6.1, giving
- and assess the impact this has on the aerosol diagnostics calculated in the previous tasks, 10.2, 10.3, and 10.4.
and were given the hint
- Remember to remove
Sec_Orgfrom the diagnostics as well as the reaction.
- Remember to remove
You should use the above python scripts provided to do this.
For a working Rose suite that has completed this task, please see
- ARCHER:
u-as292@62669 - vm:
u-as297@62632
The specific Rose changes made are:
- ARCHER: https://code.metoffice.gov.uk/trac/roses-u/changeset/62669/a/s/2/9/2/trunk
- vm: https://code.metoffice.gov.uk/trac/roses-u/changeset/62632/a/s/2/9/7/trunk
The specific Rose changes made are:
ARCHER:
Index: app/fcm_make/rose-app.conf =================================================================== --- app/fcm_make/rose-app.conf (revision 62651) +++ app/fcm_make/rose-app.conf (revision 62669) @@ -42,4 +42,4 @@ stash_version=1A timer_version=3A um_rev=vn10.9 -um_sources=branches/dev/lukeabraham/vn10.9_UKCA_Tutorial_Solns@46718 +um_sources=branches/dev/lukeabraham/vn10.9_UKCA_Tutorial_Solns@47380
These differences can be found in the file /home/ukca/Tutorial/vn10.9/worked_solutions/Task10.6/Task10.6_rose.patch on PUMA.
vm:
Index: app/fcm_make/rose-app.conf =================================================================== --- app/fcm_make/rose-app.conf (revision 62631) +++ app/fcm_make/rose-app.conf (revision 62632) @@ -42,4 +42,4 @@ stash_version=1A timer_version=3A um_rev=vn10.9 -um_sources=branches/dev/lukeabraham/vn10.9_UKCA_Tutorial_Solns@46718 +um_sources=branches/dev/lukeabraham/vn10.9_UKCA_Tutorial_Solns@47380
The specific UM changes made are:
Index: src/atmosphere/UKCA/asad_flux_dat.F90
===================================================================
--- src/atmosphere/UKCA/asad_flux_dat.F90 (revision 46718)
+++ src/atmosphere/UKCA/asad_flux_dat.F90 (revision 47380)
@@ -1287,9 +1287,9 @@
TYPE(asad_flux_defn), PARAMETER, PUBLIC :: &
ukca_tutorial_fluxes(3) = (/ &
-asad_flux_defn('RXN',50134,'B',.FALSE.,0,4, &
+asad_flux_defn('RXN',50134,'B',.FALSE.,0,3, &
(/'ALICE ','OH '/), &
-(/'BOB ','Sec_Org ',' ',' '/)), &
+(/'BOB ',' ',' ',' '/)), &
asad_flux_defn('DEP',50135,'D',.FALSE.,0,1, &
(/'ALICE ',' '/), &
(/' ',' ',' ',' '/)), &
Index: src/atmosphere/UKCA/ukca_chem_master.F90
===================================================================
--- src/atmosphere/UKCA/ukca_chem_master.F90 (revision 46718)
+++ src/atmosphere/UKCA/ukca_chem_master.F90 (revision 47380)
@@ -2156,7 +2156,7 @@
'HCHO ',1.00e-12, 0.00, 0.00, 1.00, 0.75, 0.25, 2.75, TI,0,0,107),&
ratb_t1(277,'MACRO2 ','MeOO ','HO2 ','CO ',' ',&
' ',1.00e-12, 0.00, 0.00, 1.17, 0.25, 0.00, 0.00, TI,0,0,107),&
-ratb_t1(278,'ALICE ','OH ','BOB ','Sec_Org ',' ',&
+ratb_t1(278,'ALICE ','OH ','BOB ',' ',' ',&
' ',2.70E-11, 0.00, -390.00, 0.00, 0.00, 0.00, 0.00, ST,0,0,107) /)
!----------------------------------------------------------------------
These differences can be found in the file /home/ukca/Tutorial/vn10.9/worked_solutions/Task10.6/Task10.6_code.patch on PUMA.
Sample output from this task can be found in the /work/n02/n02/ukca/Tutorial/vn10.9/sample_output/Task10.6/ directory on ARCHER, containing the following:
Task106_AOD.nc Task106_AOD_diff.nc Task106_SSA.nc Task106_SSA_diff.nc Task106_TOA.nc Task106_TOA_diff.nc atmosa.pa19810901_00 diff_AOD_rxn.py diff_SSA_rxn.py diff_TOA_rxn.py
Checklist
- ☐ Make new time and domain profiles as needed.
- ☐ Run the TidyStashTransform transform macro.
- ☐ Output your diagnostics in STASH at: um namelist Model Input and Output STASH Requests and Profiles STASH Requests.
- ☐ When adding new aerosol diagnostics, remember to output on pseudo levels if appropriate.
- ☐ Run the TidyStashTransform transform macro again for the diagnostic requests.
- ☐ Save your suite.
- ☐ In the
roses/[SUITE-ID]directory, runfcm committo commit your changes to the repository. - ☐ When modifying chemical reactions, remember to make changes for diagnostics and special cases, as well as altering
ukca_chem_master.F90. - ☐ The Iris python library can be used to process and visualise Unified Model output.
Written by Luke Abraham, Nicolas Bellouin, & Anja Schmidt 2017. With thanks to Ben Johnson.