UKCA Chemistry and Aerosol vn10.4 Tutorial 10
UKCA Chemistry and Aerosol Tutorials at vn10.4
What you will learn in this Tutorial
In this tutorial you will learn about aerosol optical depth (AOD) and how to request AOD diagnostics from UKCA. We will explain how aerosol optical properties are calculated by the RADAER module, based on the size-resolved aerosol simulated by the GLOMAP aerosol microphysics scheme.
Note: The GLOMAP aerosol tutorials require you to use u-ai071 and work from that for these tasks.
Task 10.1: What is aerosol optical depth and how do I request AOD diagnostics in UKCA?
Aerosol particles affect the Earth's radiative balance by scattering and absorbing solar radiation and, where they are large enough, can also act similarly to a greenhouse gas by absorbing outgoing terrestrial long-wave radiation.
The aerosol optical depth (AOD), sometimes referred to as aerosol optical thickness (AOT), is often used in atmospheric science as a measure of how strongly the aerosol particles within an air mass extinguish radiation (either by scattering it or absorbing it). The AOD can be thought of as a indicator of the strength of aerosol-radiation interactions at a particular wavelength.
The AOD is defined as the vertical integral of the monochromatic (single-wavelength) extinction (the sum of scattering plus absorption) through the atmospheric column. It is common also to refer to an absorption AOD which represents the integral of just the single-wavelength absorption.
The Unified Model (UM) radiation scheme divides the shortwave (SW) and longwave (LW) spectra into wavebands. To enable UKCA simulated gases or aerosols to interact with the UM radiation scheme, their optical properties need to be integrated across each of these wavebands. For aerosols, the radiation scheme requires the specific scattering and absorption coefficients, which describe the strength of aerosol scattering and absorption processes per unit aerosol mass (in m kg), and the asymmetry parameter, which describes in a simplified way the angular dependence of the scattering (dimensionless). The specific scattering and absorption coefficients, and the asymmetry parameter, are hereafter referred to as the aerosol optical properties.
Task 10.2: Understand the principles behind GLOMAP and how aerosol optical properties are derived via RADAER
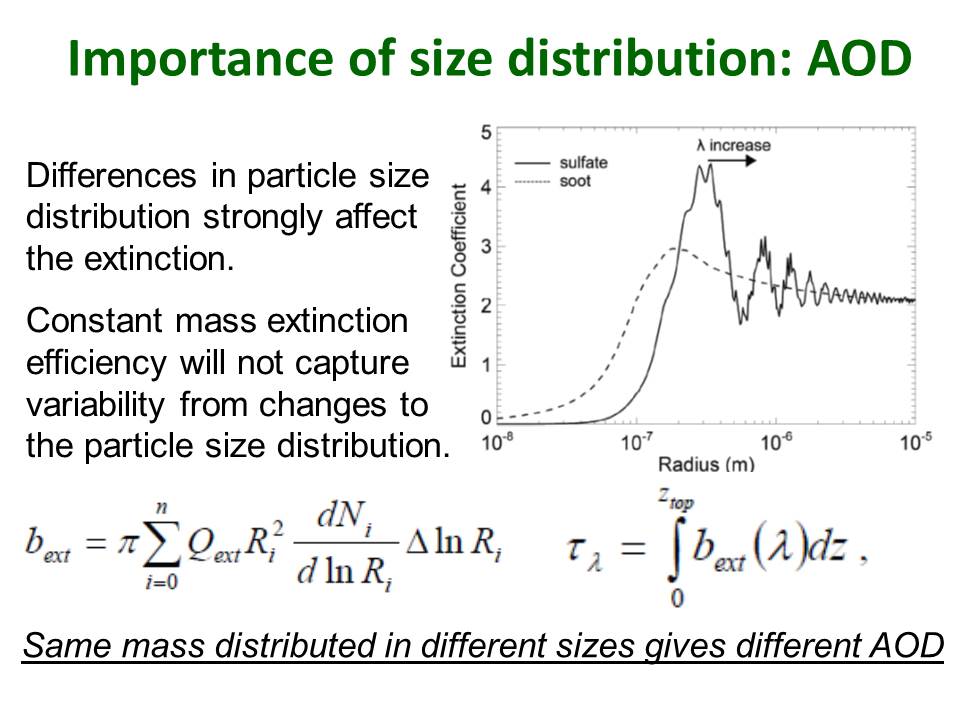
Mie theory describes the scattering and absorption of light by spherical particles. Essentially, the scattering/absorption efficiency can be parameterized in terms of two parameters: the (complex) refractive index of the particle and the Mie parameter, which describes the particle size dependence in relation to the wavelength of light under consideration.
The Figure in the slide below shows the dependence on particle size of the extinction efficiency for mid-visible (550nm, green) light for two different types of particle (refractive index).
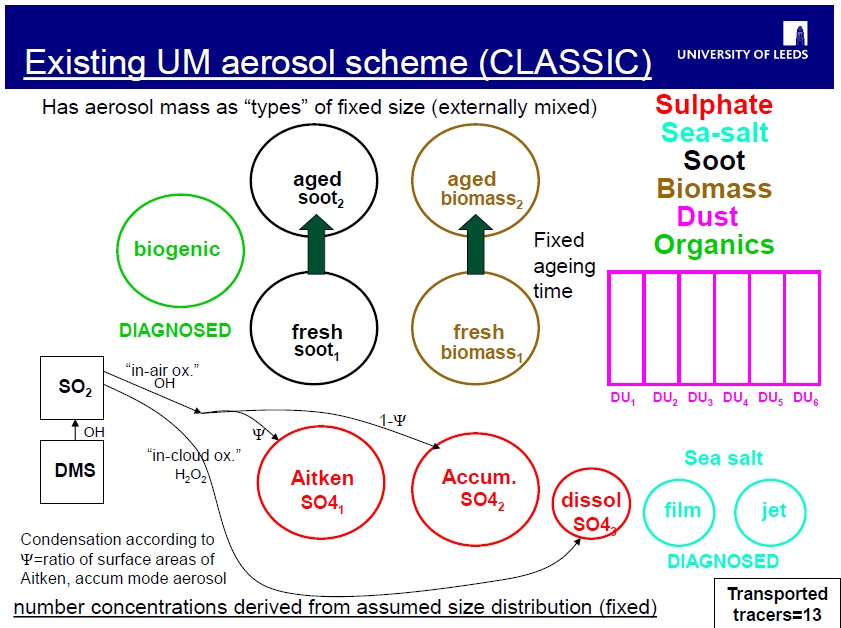
The aerosol scheme used for UK climate model simulations in CMIP5 was a simplified mass-based scheme Coupled Large-scale Aerosol Simulator for Studies In Climate (CLASSIC, e.g. Bellouin et al., 2007). Although CLASSIC is simpler than the microphysical GLOMAP scheme included in UKCA, it does have a comprehensive representation of the main aerosol sources, tracking up to eight tropospheric aerosol species: ammonium sulphate, mineral dust, fossil fuel black carbon (soot), fossil fuel organic carbon, biomass burning aerosols, sea-salt, secondary organic (also called biogenic), and ammonium nitrate aerosols. The first 5 of these are treated interactively in terms of their mass, but prescribe globally and temporally uniform values for their size distribution and refractive index. Some variation in aerosol properties was resolved by having fresh and aged sub-types for some of these types, which then allow size and refractive index to vary from values near-source (fresh) to remote (aged).
With this simplified approach, optical properties for each aerosol type are derived based on Mie calculations using the uniform size distribution and refractive index for each type with water uptake based on a fixed hygroscopic growth factor. For all three properties the constants used are based on average values observed in air masses dominated by the particular type.
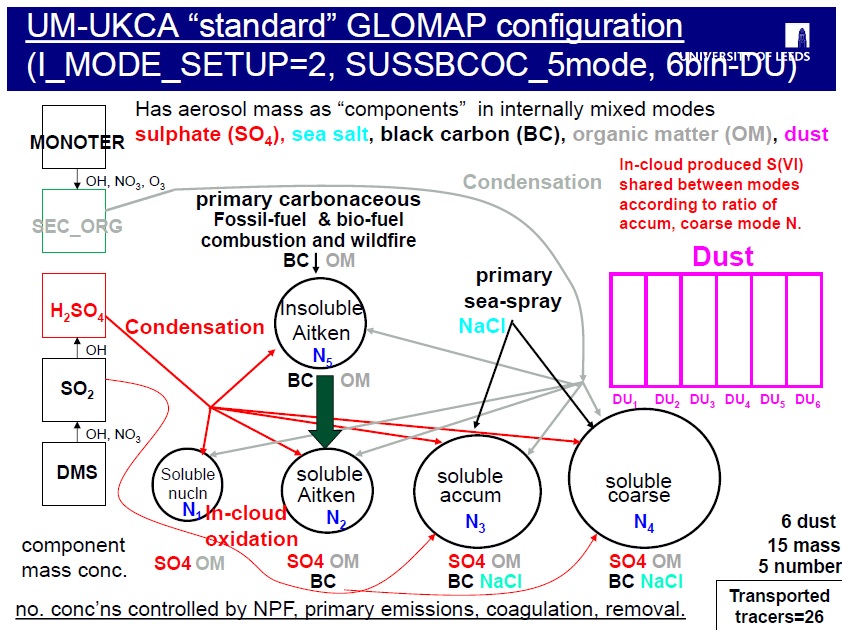
The GLOMAP-mode aerosol scheme in UKCA differs in that it simulates the variation of size-resolved aerosol properties, as they evolve due to microphysical processes such as new particle formation, coagulation, condensation (gas-to-particle-transfer) and cloud processing.
Whereas CLASSIC simulated only the mass of several aerosol types, the transported tracers in GLOMAP are particle number and mass in different size classes spanning the particle size range from 3nm up to around 20 microns dry diameter.
Processes such as condensation and aqueous sulphate production grow particles to larger sizes by increasing the mass in a size class while conserving particle number.
As was as microphysical processes, GLOMAP also includes size-resolved representations of primary emissions (e.g. of sea-salt, dust and carbonaceous particles), and also has size-resolved removal processes including particle dry deposition, sedimentation, nucleation scavenging (rainout) and impaction scavenging (washout).
GLOMAP therefore simulates the evolution of particle number and composition across the size spectrum over several different chemical constituents or components (e.g. sea-salt, sulphate, black carbon) as determined by the aerosol processes included.
The original version of GLOMAP (known as GLOMAP-bin) uses a two-moment sectional aerosol dynamics approach, with typically 20 bins spanning the size spectrum, but when tracking several aerosol types, becomes too expensive for running multi-decadal integrations as required in a climate model. For UKCA, a new computationally cheaper version of GLOMAP was developed (GLOMAP-mode), which has the same process representations, but using log-normal modes as its size classes. Each size mode in GLOMAP-mode covers one of four size ranges (nucleation, Aitken, accumulation, coarse), and additionally distinguishing between soluble and insoluble modes.
GLOMAP has been developed to follow a flexible multi-component approach, with the same code able to be run with different levels of composition/size sophistication with FORTRAN-90 modules providing alternative aerosol mode set-up arrays. The GLOMAP-mode aerosol scheme in UKCA not only simulates the dry aerosol mass, but also the mass of water attached to the aerosol, and the aerosol number concentrations.
The calculation of aerosol optical properties from GLOMAP is carried out by the RADAER module included in UKCA.
Compared to CLASSIC, the GLOMAP-mode scheme introduces three important changes which are relevant for simulated aerosol-radiation interactions:
- The mean radius of each size class (mode) varies in time and space as determined by transport and aerosol processes.
- There is a refractive index for each size class (mode) which varies according to its internally-mixed composition.
- The aerosol water taken up into each soluble mode varies interactively, consistently with particle composition.
RADAER relies on pre-computed look-up tables of monochromatic optical properties, covering all realistic combinations of modal radii and refractive indices. At runtime, remaining tasks are:
- To compute the modal refractive index out of the simulated chemical composition of each mode.
- To obtain the monochromatic properties from the look-up tables at selected wavelengths within each SW and LW waveband.
- To integrate across each waveband to obtain the waveband-averaged optical properties, which can be used by the radiation code.
As well as providing the waveband-averaged scattering, extinction and asymmetry parameters from GLOMAP, RADAER also diagnoses monochromatic Aerosol Optical Depth (AOD) for each of the GLOMAP modes.
Task 10.3: Output daily-mean Aerosol Optical Depths from your UKCA model run
In this task you will update your copy of the UKCA tutorial suite (u-ai071) adding
extra STASH requests to output daily-mean AODs from GLOMAP.
UM-UKCA diagnoses the AOD from each of the GLOMAP modes as a separate STASH item.
All of the GLOMAP AOD diagnostics are contained with STASH section 2 (long-wave radiation) in item numbers close to those used for the AOD diagnostics from CLASSIC.
Section 2 items 300-305 contain the AOD for the Aitken-soluble, accumulation-soluble, coarse-soluble, Aitken-insoluble, accumulation-insoluble and coarse-insoluble modes respectively.
The dimensions for each of these STASH items is 2D global in longitude and latitude but there is also a third dimension containing 6 pseudo levels for the 6 monochromatic AODs stored by the model (at 0.38, 0.44, 0.55, 0.67, 0.87, 1.02 micron wavelengths).
Note that there is no nucleation mode AOD as those particles are too small to significantly scatter or absorb radiation at these wavelengths.
There are separate "stratospheric AOD" diagnostics in section 2 items 251 to 256 which can also be requested to store the AOD in levels above the tropopause.
Note there are also absorption AOD in these 6 modes can usually be requested via STASH section 2 items 240 to 245, in the job here these are not available in the UKCA tutorial suite.
To output AODs from GLOMAP, you need to add in some extra STASH requests for the section 2 items 300 to 305.
To output as a daily-mean select the usage profile "UPK" to output to the .pk files and the time profile "TDAYM" for daily-means. For AOD STASH requests, you need to select the domain profile DIAGAOT to output over the longitude, latitude and 6 wavelength pseudo-levels.
In the next task (Task 11), you will change the GLOMAP aerosol setup to have two different OM components in each size mode, to allow to track two different types of particulate OM. In anticipation of that task, as well as the AOD for each mode, we'll also request the mass mixing ratio of OM in each mode.
Section 34 items 106, 110, 116, 121 and 126 contain the OM mmr's in the Aitken-soluble, accumulation-soluble, coarse-soluble, Aitken-insoluble and nucleation-soluble modes respectively.
When requesting the OM mmr's, use the DALLTH domain profile, as those variables are on the full 3D model grid.
After doing that you will have an equivalent version of the UKCA tutorial job with a number of additional daily-mean fields requested.
Since you specified the UPK usage profile in the job, these fields will be output to the so-called .pk files.
These can be found in your /work/n02/n02/ directory on ARCHER. You should have a file like jobida.pk19880901 for the daily-mean fields for the 1 day that the job was set to run for.
In the first instance you should use xconv to open the file although you can use convsh or other tools to extract required fields from the .pk file and then manipulate them in IDL or python.
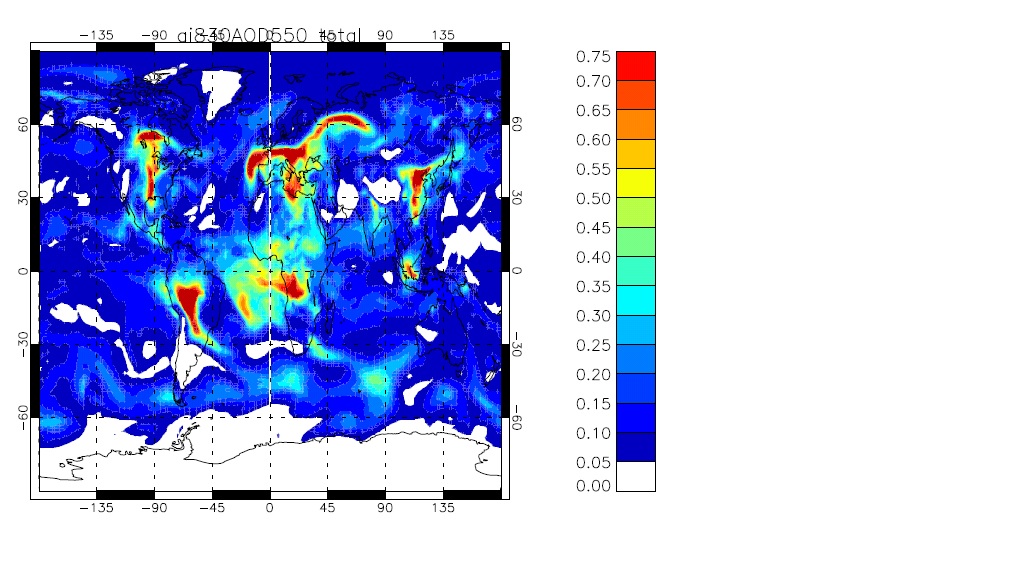
As an example I have put below a jpeg showing a global map produced in IDL of the daily-mean AOD at 550nm (total
over all modes) for 1st September 1988 from the UKCA tutorial suite (worked solution u-ai830).
In preparation for the next task, you should also re-run the job adding also daily-mean STASH requests for the following mass mixing ratios (mmr's) for the Organic Matter (OM) in each mode (34106, 34110, 34116, 34121 and 34126).
Once you have re-run the job with these extra fields available in the .pk file, you can use the cdo operator cdo add to sum up each of the OM mmrs to give a total OM mmr in a separate netCDF file.
You should now cd into your $HOME/roses/[SUITE-ID] directory and fcm commit your changes.
Worked Solution
A worked solution to Task 10.3 is job u-ai830.
Your .pk file will contain the following fields:
0 : 192 144 6 1 Unspecified 1 : 192 144 6 1 Unspecified 2 : 192 144 6 1 Unspecified 3 : 192 144 6 1 Unspecified 4 : 192 144 6 1 Unspecified 5 : 192 144 6 1 Unspecified 6 : 192 144 6 1 Unspecified 7 : 192 144 6 1 Unspecified 8 : 192 144 6 1 Unspecified 9 : 192 144 6 1 Unspecified 10 : 192 144 6 1 Unspecified 11 : 192 144 6 1 Unspecified 12 : 192 144 85 1 AITKEN MODE (SOLUBLE) OC MMR 13 : 192 144 85 1 ACCUMULATION MODE (SOL) OC MMR 14 : 192 144 85 1 COARSE MODE (SOLUBLE) OC MMR 15 : 192 144 85 1 AITKEN MODE (INSOLUBLE) OC MMR 16 : 192 144 85 1 NUCLEATION MODE (SOLUBLE) OC MMR
You can use cdo as so to add the 5 separate netCDF files containing the OC mass-mixing ratios. To do this you must first
module load cdo
to give you access to the command. You can extract the required fields from the .pk file using Xconv or convsh. An example convsh script is below:
#! /usr/bin/env convsh # This file is part of the UKCA Tutorials: # http://www.ukca.ac.uk/wiki/index.php/UKCA_Chemistry_and_Aerosol_Tutorials_at_vn10.4 # Copyright (C) 2015 University of Cambridge # This is free software: you can redistribute it and/or modify it under the # terms of the GNU Lesser General Public License as published by the Free Software # Foundation, either version 3 of the License, or (at your option) any later # version. # It is distributed in the hope that it will be useful, but WITHOUT ANY # WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A # PARTICULAR PURPOSE. See the GNU Lesser General Public License for more details. # You find a copy of the GNU Lesser General Public License at <http://www.gnu.org/licenses/>. # Written by N. Luke Abraham 2017-01-06 <nla27@cam.ac.uk> # you should change the input file name here readfile 0 ai922a.pk19880901 # You may need to change the field numbers (first column in Xconv or when using list_fields) # You should also change the prepended suite-id # These should match these names: # AITKEN MODE (SOLUBLE) OC MMR writefile netcdf ai922_m01s34i106.nc 12 # ACCUMULATION MODE (SOL) OC MMR writefile netcdf ai922_m01s34i110.nc 13 # COARSE MODE (SOLUBLE) OC MMR writefile netcdf ai922_m01s34i116.nc 14 # AITKEN MODE (INSOLUBLE) OC MMR writefile netcdf ai922_m01s34i121.nc 15 # NUCLEATION MODE (SOLUBLE) OC MMR writefile netcdf ai922_m01s34i126.nc 16
These can then be combined using the following cdo command:
cdo add ai922_m01s34i106.nc -add ai922_m01s34i110.nc -add ai922_m01s34i116.nc -add ai922_m01s34i121.nc ai922_m01s34i126.nc ai922_OCmmr_cdo.nc
When opening the output file (ai922_OCmmr_cdo.nc in the example above) the variable name is taken from the first file in the list.
It is also possible to use Iris to extract and combine these fields - an example script showing how to do this is below:
#!/usr/bin/env python # This file is part of the UKCA Tutorials: # http://www.ukca.ac.uk/wiki/index.php/UKCA_Chemistry_and_Aerosol_Tutorials_at_vn10.4 # Copyright (C) 2015 University of Cambridge # This is free software: you can redistribute it and/or modify it under the # terms of the GNU Lesser General Public License as published by the Free Software # Foundation, either version 3 of the License, or (at your option) any later # version. # It is distributed in the hope that it will be useful, but WITHOUT ANY # WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A # PARTICULAR PURPOSE. See the GNU Lesser General Public License for more details. # You find a copy of the GNU Lesser General Public License at <http://www.gnu.org/licenses/>. # Written by N. Luke Abraham 2017-01-06 <nla27@cam.ac.uk> # preamble import iris # load all OC components oc=iris.load('./ai922a.pk19880901', [iris.AttributeConstraint(STASH='m01s34i106'), iris.AttributeConstraint(STASH='m01s34i110'), iris.AttributeConstraint(STASH='m01s34i116'), iris.AttributeConstraint(STASH='m01s34i121'), iris.AttributeConstraint(STASH='m01s34i126')]) # make cube to store total OC ocsum=oc[0].copy() # add-up components ocsum.data=oc[0].data+oc[1].data+oc[2].data+oc[3].data+oc[4].data # rename ocsum.rename('mass_fraction_of_total_particulate_organic_matter_in_air') # remove unlimited dimension when writing to netCDF iris.FUTURE.netcdf_no_unlimited=True # output to netCDF iris.save(ocsum,'ai922_OCmmr_iris.nc',netcdf_format='NETCDF3_CLASSIC')
It is also possible to use Iris to calculate the 0.55 micron AOD - an example script to do this is below:
#!/usr/bin/env python # This file is part of the UKCA Tutorials: # http://www.ukca.ac.uk/wiki/index.php/UKCA_Chemistry_and_Aerosol_Tutorials_at_vn10.4 # Copyright (C) 2015 University of Cambridge # This is free software: you can redistribute it and/or modify it under the # terms of the GNU Lesser General Public License as published by the Free Software # Foundation, either version 3 of the License, or (at your option) any later # version. # It is distributed in the hope that it will be useful, but WITHOUT ANY # WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A # PARTICULAR PURPOSE. See the GNU Lesser General Public License for more details. # You find a copy of the GNU Lesser General Public License at <http://www.gnu.org/licenses/>. # Written by N. Luke Abraham 2017-01-06 <nla27@cam.ac.uk> # preamble import iris # load all AOD components at 0.55 micron aod=iris.load('./ai922a.pk19880901', [iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i300'), iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i301'), iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i302'), iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i303'), iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i304'), iris.Constraint(pseudo_level=3) & iris.AttributeConstraint(STASH='m01s02i305')]) # make cube to store total AOD aodsum=aod[0].copy() # add-up components aodsum.data=aod[0].data+aod[1].data+aod[2].data+aod[3].data+aod[4].data+aod[5].data # rename aodsum.rename('atmosphere_optical_thickness_due_to_aerosol') # remove unlimited dimension when writing to netCDF iris.FUTURE.netcdf_no_unlimited=True # output to netCDF iris.save(aodsum,'ai922_AOD_iris.nc',netcdf_format='NETCDF3_CLASSIC')
Example output can be found on ARCHER in the directory
/work/n02/n02/ukca/Tutorial/vn10.4/sample_output/Task10.3
The Rose-specific changes can be found in the file
/home/ukca/Tutorial/worked_solutions/Task10.3/task10.3.rose.diff
on PUMA
Index: app/um/rose-app.conf =================================================================== --- app/um/rose-app.conf (revision 28511) +++ app/um/rose-app.conf (revision 28536) @@ -4862,6 +4862,14 @@ tim_name='TDMPMN' use_name='UPMEAN' +[namelist:streq(1cdb7a57)] +dom_name='DALLTH' +isec=34 +item=106 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(1cedac0b)] dom_name='DIAG' isec=3 @@ -4886,6 +4894,14 @@ tim_name='TDAYM' use_name='UPD' +[namelist:streq(1d51d9e9)] +dom_name='DALLTH' +isec=34 +item=116 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(1d882b73)] dom_name='DIAG' isec=1 @@ -4934,6 +4950,14 @@ tim_name='T90DAY' use_name='UPI' +[namelist:streq(1fa7073a)] +dom_name='DALLTH' +isec=34 +item=126 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(1fbc22ae)] dom_name='DP17' isec=30 @@ -6454,6 +6478,14 @@ tim_name='TMPMN18' use_name='UPMEAN' +[namelist:streq(5e6e0742)] +dom_name='DALLTH' +isec=34 +item=110 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(5f297582)] dom_name='DIAG' isec=30 @@ -6606,6 +6638,14 @@ tim_name='TRADDM' use_name='UPMEAN' +[namelist:streq(67a9602e)] +dom_name='DIAGAOT' +isec=2 +item=254 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(67e9519f)] dom_name='DALLTH' isec=1 @@ -6686,6 +6726,14 @@ tim_name='TDMPMN' use_name='UPMEAN' +[namelist:streq(6b285660)] +dom_name='DIAGAOT' +isec=2 +item=255 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(6b37b902)] dom_name='DIAG' isec=1 @@ -6782,6 +6830,14 @@ tim_name='TDMPMN' use_name='UPMEAN' +[namelist:streq(6ebee303)] +dom_name='DIAGAOT' +isec=2 +item=252 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(6ec24c21)] dom_name='DTROP' isec=5 @@ -6902,6 +6958,14 @@ tim_name='TDMPMN' use_name='UPMEAN' +[namelist:streq(74c20937)] +dom_name='DIAGAOT' +isec=2 +item=256 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(75413c65)] dom_name='DTROP' isec=5 @@ -7550,6 +7614,14 @@ tim_name='TALLTS' use_name='UPUKCA' +[namelist:streq(8f865257)] +dom_name='DALLTH' +isec=34 +item=121 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(8fc8c95a)] dom_name='DNOGWTH' isec=6 @@ -7830,6 +7902,14 @@ tim_name='T6HDMPM' use_name='UPMEAN' +[namelist:streq(9f3b2872)] +dom_name='DIAGAOT' +isec=2 +item=253 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(9f60f084)] dom_name='DIAG' isec=1 @@ -8262,6 +8342,14 @@ tim_name='TDMPMN' use_name='UPMEAN' +[namelist:streq(b45df417)] +dom_name='DIAGAOT' +isec=2 +item=300 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(b4656f70)] dom_name='DIAG' isec=4 @@ -8438,6 +8526,14 @@ tim_name='TDAYM' use_name='UPD' +[namelist:streq(bb6af665)] +dom_name='DIAGAOT' +isec=2 +item=251 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(bccca6bc)] dom_name='DIAG' isec=5 @@ -8678,6 +8774,14 @@ tim_name='TMPMN00' use_name='UPMEAN' +[namelist:streq(c49b9649)] +dom_name='DIAGAOT' +isec=2 +item=303 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(c5a142ea)] dom_name='DIAG' isec=3 @@ -9390,6 +9494,14 @@ tim_name='TDMPMN' use_name='UPMEAN' +[namelist:streq(e3f9fc46)] +dom_name='DIAGAOT' +isec=2 +item=305 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(e40c8701)] dom_name='DIAG' isec=5 @@ -9494,6 +9606,14 @@ tim_name='TDMPMN' use_name='UPMEAN' +[namelist:streq(e924a4a3)] +dom_name='DIAGAOT' +isec=2 +item=304 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(e92db8ee)] dom_name='DALLTH' isec=50 @@ -9646,6 +9766,14 @@ tim_name='TDMPMN' use_name='UPMEAN' +[namelist:streq(f1347f13)] +dom_name='DIAGAOT' +isec=2 +item=302 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(f14e5386)] dom_name='DIAG' isec=3 @@ -9790,6 +9918,14 @@ tim_name='TDMPMN' use_name='UPMEAN' +[namelist:streq(f5e063cb)] +dom_name='DIAGAOT' +isec=2 +item=301 +package= +tim_name='TDAYM' +use_name='UPK' + [namelist:streq(f5e615d1)] dom_name='DIAG' isec=5 Index: rose-suite.conf =================================================================== --- rose-suite.conf (revision 28511) +++ rose-suite.conf (revision 28536) @@ -1,7 +1,7 @@ [jinja2:suite.rc] !!ACCOUNT_USR='foundation' ANCIL_OPT_KEYS= -ARCHER_GROUP='n02-chem' +ARCHER_GROUP='n02-training' ARCHER_QUEUE='short' !!ARCH_LOG=true !!ARCH_WALL=true @@ -21,7 +21,7 @@ !!HOST_XC40='xc' HOUSEKEEP=true !!HPC_QUEUE='normal' -HPC_USER='luke' +HPC_USER='ncastr01' HYPTHR_ATM=1 HYPTHR_RCF=1 IOS_NPROC=0
Written by Graham Mann 2016