Difference between revisions of "UKCA Chemistry and Aerosol UMvn13.0 Tutorial 3"
| Line 7: | Line 7: | ||
| Time to Complete || '''1-2 hours''' |
| Time to Complete || '''1-2 hours''' |
||
|- |
|- |
||
| − | | Video instructions || |
+ | | Video instructions || [https://www.youtube.com/watch?v=n3yE-oYy-gU&t=1726s Walkthrough (YouTube)] |
|} |
|} |
||
Latest revision as of 13:46, 13 May 2023
UKCA Chemistry and Aerosol Tutorials at UMvn13.0
| Difficulty | MEDIUM |
| Time to Complete | 1-2 hours |
| Video instructions | Walkthrough (YouTube) |
Remember to run mosrs-cache-password.
What you will do in this tutorial
In this tutorial you will learn how to make a branch to allow you to include UKCA code changes within the Box Model. You will then include 2 new chemical tracers, ALICE and BOB.
To be able to make UKCA code changes you will need to make a work with the UKCA code repository on the Met Office Science Repository Service (MOSRS), located here
Working with the UKCA repository
Making a ticket
To work with the UKCA code you will need to first make a new "ticket" on the UKCA trac pages on MOSRS, located here:
You should ensure that:
- The Milestone is set to Not for builds. If you were developing a code change for the trunk then this would be set to the code version you wanted the change to be available from.
- Once the ticket is created you click Modify Ticket and then select Start work and then Submit changes. This will make the ticket active and owned by you.
An example ticket can be seen at
The ticket is useful to be able to keep a log of your progress with the ticket as well as being able to hold trac wiki pages that can be used to present results and documentation.
In these tutorials we will have tickets and branches for both the UKCA and UM, so you will need to keep track of both.
Make a branch
Once you have a ticket you are able to make a branch on the UKCA repository. You should do this in your terminal
fcm branch-create --type dev -k ticket_number your_branch_name fcm:ukca.x_tr@um13.0
| Setting | Description |
|---|---|
| ticket_number | This is the number of the ticket that you have just created, e.g. 33 etc. |
| your_branch_name | This is a descriptive name for your branch, e.g. ukca_tutorials. You should not also include a revision/version number here. |
| REV i.e. um13.0 |
This is the trunk revision we are branching from. For these tutorials it is um13.0. |
Once you have run this command you will be placed into a text editor (as when you copied your Rose suite) to provide a commit message, e.g.
--Add your commit message ABOVE - do not alter this line or those below-- --FCM message (will be inserted automatically)-- #33: Created /main/branches/dev/lukeabraham/um13.0_ukca_tutorial_solutions from /main/trunk@264. --Change summary (not part of commit message)-- A https://code.metoffice.gov.uk/svn/ukca/main/branches/dev/lukeabraham/um13.0_ukca_tutorial_solutions
You should put a descriptive commit message, e.g. something like
#33 lukeabraham creating branch for UKCA tutorials
The #33 will resolve as a link on the trac webpages, allowing you to easily find the ticket, and providing your name is useful to keep track of who made changes. Now you should save and quit the text editor. In the terminal, you will then be presented with
[info] Source: https://code.metoffice.gov.uk/svn/ukca/main/trunk@264 (264) [info] vim: starting commit message editor... Change summary: -------------------------------------------------------------------------------- A https://code.metoffice.gov.uk/svn/ukca/main/branches/dev/lukeabraham/um13.0_ukca_tutorial_solutions -------------------------------------------------------------------------------- Commit message is as follows: -------------------------------------------------------------------------------- #33 lukeabraham creating branch for UKCA tutorials #33: Created /main/branches/dev/lukeabraham/um13.0_ukca_tutorial_solutions from /main/trunk@264. -------------------------------------------------------------------------------- Create the branch? Enter "y" or "n" (or just press <return> for "n")
You should now type y and press return. This will create the branch.
Now change directory to one where you want to copy your branch to, e.g. cd and then return to take you back to your home directory. You should then select the full URL of the branch and check it out using the command
fcm co
e.g.
fcm co https://code.metoffice.gov.uk/svn/ukca/main/branches/dev/[YOUR MOSRS USERNAME]/um13.0_[YOUR BRANCH NAME]
You can then cd into this directory. The UKCA source code is held within the src directory. The code is organised in the following directory structure, with a number of Fortran source files in each directory and sub-directory
control control/core control/core/diagnostics control/core/interface control/core/misc control/core/top_level control/legacy science science/core science/core/aerosols science/core/aerosols/deposition science/core/aerosols/glomap science/core/aerosols/activation science/core/chemistry science/core/chemistry/deposition science/core/chemistry/asad science/core/emissions science/radaer science/photolysis science/photolysis/stratospheric science/photolysis/stratospheric/photolib science/photolysis/fastjx science/photolysis/tropospheric science/glomap_clim
Use your branch in your suite
Although we have not made any code changes yet, we will include our branch in our Rose suite now so that the changes will get picked-up the next time we run. To do this, go to the fcm_make env Sources panel. You will need to click the green plus symbol (+ for the ukca_sources to add a new line, and then copy and paste the path to the working directory of your branch, e.g.
/home/vagrant/um13.0_ukca_tutorial_solutions
It is also possible to specify the repository location on the MOSRS using
branches/dev/[YOUR MOSRS USERNAME]/um13.0_[YOUR BRANCH NAME]@[REV]
where the @[REV] (which is optional) is the revision of the latest version of the branch, e.g. 471. However, this requires you to commit your code before it is picked-up by the model, and so for code development it is often easier to point to a working copy.
The change to the suite is now, e.g.
Index: app/fcm_make/rose-app.conf =================================================================== --- app/fcm_make/rose-app.conf (revision 236496) +++ app/fcm_make/rose-app.conf (working copy) @@ -30,6 +30,6 @@ thread_utils=false timer_version=3A ukca_rev=$BASE_UKCA_REV -ukca_sources=/home/vagrant/um13.0_ukca_box +ukca_sources=/home/vagrant/um13.0_ukca_box /home/vagrant/um13.0_ukca_tutorial_solutions um_rev=$BASE_UM_REV um_sources=
So it is uses 2 branches, the new one and the original /home/vagrant/um13.0_ukca_box branch that contains the control routines for the box model. You need both branches for the remainder of these tutorials.
Include your new chemical tracers
Before we can consider any chemical reactions with new species, we first need to include these as UKCA chemical tracers. This is done in 4 places:
- In the ukca_chem_master.F90 routine, to add them to the master species list
- In the ukca_config_defs_mod.F90 routine, to tell the relevant chemical mechanism how many chemical tracers it has
- In the ukca_constants.F90 routine, to define conversion factors for each species from mass-mixing ratio to mole fraction
- In the ukca_cspecies.F90 routine, to assign these conversion factors to the correct place in the master list of conversion factors
Task 3.1: Include ALICE and BOB
TASK 3.1: Include ALICE and BOB as chemical tracers for both the StratTrop and CRI-Strat schemes. You should set the conversion factor for each of these to 1.0.
src/science/core/chemistry/ukca_chem_master.F90
In this file the list of species used in chemistry is defined by the array chch_defs_master. The list of species is held in a derived type, of the format:
chch_t1( N,'SPECIES ', X,'TYPE ','FAMILY ', D, W, SCHEME, QUALIFIER, DISQUALIFIER, VN), &
Where:
- N is an integer. The value should be incremented for each new species that is added so that each species has a unique number, but there can be several entries for each species.
- 'SPECIES ' is the name of the species, e.g. 'O3 '
- X is number of odd atoms. This field is not currently used so please set it to 1.
- 'TYPE ' describes whether the species is
- a tracer ('TR ')
- an RO2 tracer ('OO ')
- a steady-state species ('SS ')
- a constant ('CT ')
- a constant field ('CF ').
- For SS, CT, and CF, special code will need to be added.
- 'FAMILY ' is the family that the species belongs to. This field is not currently used.
- D is 1 if the species is dry-deposited, and 0 otherwise.
- W is 1 if the species is wet-deposited, and 0 otherwise.
- SCHEME defines the chemistry scheme used, e.g.:
- st = Stratosphere-Troposphere scheme
- t = Troposphere scheme
- s = Stratosphere scheme
- r = RAQ scheme
- ol = Offline-oxidants scheme
- ti = Troposphere-Isoprene scheme
- cs = CRI-Strat scheme
- If the same species is used in multiple schemes these are added, e.g. st+s+ti+cs etc.
- QUALIFIER (and DISQUALIFIER) defines lines to add (or remove) on top of the base chemistry scheme depending on if certain additional options have been chosen, e.g.
- 0 = no qualifier/disqualifer used
- a = Aerosol chemistry additions required to run GLOMAP-mode
- th = Tropospheric heterogeneous reactions
- hp = Heterogeneous PSC chemistry
- es = Extended stratospheric reactions
- ci = use a 3-D CO2 tracer
- eh = Extended heterogeneous chemistry
- rn = RO2 a non-transported prognostic (StratTrop and CRI-Strat chemistry only)
- rp = RO2 permutation chemistry (StratTrop and CRI-Strat chemistry only)
- cs2 = used for changes from CRI-Strat version 1 to CRI-Strat version 2 chemistry
- ce = used for emissions-driven CH4
- rm = used to remove reactions from older mechanisms
- As with the SCHEME codes, these are also additive if required, e.g. a+th etc.
- VN gives the model version the addition is added at. This is future functionality that will allow for multiple rates to be included in the source-code. The current standard version is 111, but for these tutorials we are using 121, which contains the latest rate updates.
Therefore to add-in the ALICE and BOB species, we should insert lines similar to this
! 284 chch_t1(284,'ALICE ',1,'TR ',' ',0,0,st+cs,0,0,131), & ! 285 chch_t1(285,'BOB ',1,'TR ',' ',0,0,st+cs,0,0,131)
into chch_defs_master, remembering to also increment the size of this array, given by n_chch_master.
Note that the version has also changed for these species.
src/control/core/top_level/ukca_config_defs_mod.F90
In ukca_config_defs_mod.F90 you will find code blocks which set values for various parameters depending on the scheme used. In this example you will need to scroll down to the sections that define the StratTrop and CRI-Stratchemistry schemes. When adding tracers you will need to increment the value of n_chem_tracers.
For StratTrop this is located in the Stratospheric Chemistry section and is referenced by using the ukca_config%l_ukca_strattrop logical. For CRI-Strat this is located in the CRI-Strat and CRI-Strat 2 chemistry section controlled by ukca_config%l_ukca_cristrat logical. Note also for CRI-Strat there is an additional check on the chemistry version number used, as if this is greater than 119 we are using CRI-Strat version 2.
The IF blocks test against ukca_config%l_ukca_achem (which determines whether or not you require the additional chemistry used to drive the GLOMAP-mode aerosol scheme). You should increment the value of n_chem_tracers by the number of chemical tracers that you are adding (i.e. 2) in both sections of these IF blocks, as this tracer is not defined to be for aerosol chemistry only. If your additional tracers were only aerosol chemistry additions then you would increment the value of n_aero_tracers. Note that as we are using a chemical version of 131 we only need to increment the value for CRI-Strat2.
If you are adding to a different chemistry scheme then you will need to make these changes accordingly
src/science/core/chemistry/ukca_constants.F90
The unit of the tracers is kg(species)/kg(air) (i.e. mass mixing ratio, or mmr), but inside UKCA these species are converted to mole fraction or volume mixing ratio (vmr). To enable UKCA to do this you will need to add the conversion factor(s) for your new tracer(s) into the code. This is done in the ukca_constants.F90 module.
Open this file and add the required conversion factor(s). The naming convention for these is:
- m_species for the molecular mass of the new species in g/mol
- c_species for the converson factor from vmr to mmr
The c_species conversion factor calculated as m_species/M_air, where M_air=28.97). In actuality, only the c_species listing is always required, although you may need to add the M_species value later if you are emitting into the new species that you are adding.
For these new species we can add the following blocks, as we have been told to use a conversion factor of 1:
REAL, PARAMETER :: c_alice = 1.000 REAL, PARAMETER :: c_bob = 1.000
src/science/core/chemistry/ukca_cspecies.F90
After you have added in the c_species conversion factors, you will need to tell UKCA to use them for your species. To do this you need to edit the ukca_cspecies.F90 module, which contains code which constructs the c_species array of conversion factors for the advected tracers. This contains a subroutine called ukca_calc_cspecies which has a long block of code that you need to edit to add entries like this:
WHERE (advt == 'ALICE ') c_species = c_alice WHERE (advt == 'BOB ') c_species = c_bob
The advt array is automatically generated by UKCA at run-time from the chch_defs_master chemistry scheme definition you edited earlier, so your new tracer(s) will exist within it. You need to add in a new line for each of your tracers which sets the value of the c_species array to your individual c_species parameter. Add the line(s) in at the end of the block.
As the c_species conversion factors are defined in the ukca_constants module you will need to explicitly add them to the
USE ukca_constants, ONLY: ...
statement at the top of the module.
Edit your Rose Suite
As we have used chemistry version 131 for these new species, we will need to edit the value of i_ukca_chem_version to be 131. You should now run your suite and see what happens with ALICE and BOB.
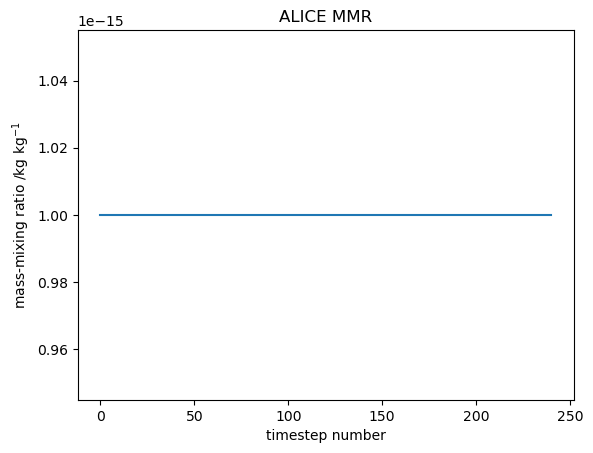
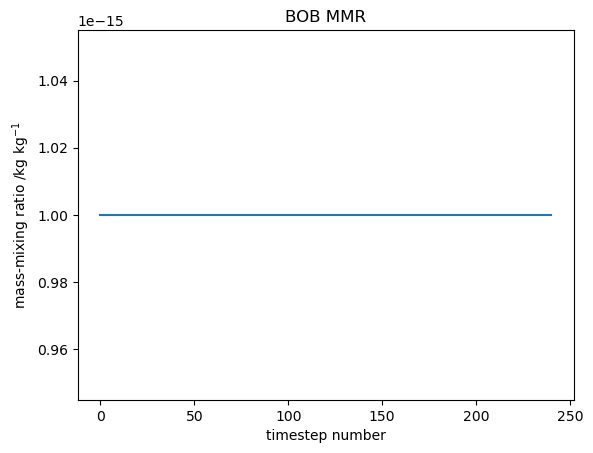
Note that initial conditions of ALICE and BOB do not need to be specified at this stage as any species that does not have a concentration explicitly given in tracer_in_filename uses the value given by tracer_nullval, currently set to 1e-15.
Worked Solution to Task 3.1
As there is no chemistry occurring for species, and they are initialised to very small values, they will maintain there values throughout the simulation.
The changes to Rose are:
- https://code.metoffice.gov.uk/trac/roses-u/changeset/236512/c/q/7/8/2/trunk
- https://code.metoffice.gov.uk/trac/roses-u/changeset/236520/c/q/7/8/2/trunk
Index: app/ukca/rose-app.conf =================================================================== --- app/ukca/rose-app.conf (revision 236496) +++ app/ukca/rose-app.conf (revision 236520) @@ -481,7 +481,7 @@ !!i_mode_setup=2 !!i_ukca_activation_scheme=0 i_ukca_chem=59 -i_ukca_chem_version=121 +i_ukca_chem_version=131 i_ukca_conserve_method=3 !!i_ukca_dms_flux=1 !!i_ukca_hetconfig=0 Index: app/fcm_make/rose-app.conf =================================================================== --- app/fcm_make/rose-app.conf (revision 236496) +++ app/fcm_make/rose-app.conf (revision 236520) @@ -30,6 +30,6 @@ thread_utils=false timer_version=3A ukca_rev=$BASE_UKCA_REV -ukca_sources=/home/vagrant/um13.0_ukca_box +ukca_sources=/home/vagrant/um13.0_ukca_box /home/vagrant/um13.0_ukca_tutorial_solutions um_rev=$BASE_UM_REV um_sources=
These differences can be found here:
Tutorials/UMvn13.0/worked_solutions/Task03.1/Task03.1_rose.patch
The changes to UKCA are:
Index: src/control/core/top_level/ukca_config_defs_mod.F90
===================================================================
--- src/control/core/top_level/ukca_config_defs_mod.F90 (revision 471)
+++ src/control/core/top_level/ukca_config_defs_mod.F90 (revision 475)
@@ -315,7 +315,7 @@
['NO ','CH4 ','CO ','HCHO ', &
'C2H6 ','C3H8 ','Me2CO ','MeCHO ', &
'C5H8 ', nm_meoh_em ,'NO_aircrft']
- n_chem_tracers = 71 ! No chem tracers
+ n_chem_tracers = 73 ! No chem tracers
nr_therm = 220 ! thermal reactions
nr_phot = 55 ! photolytic (ATA)
@@ -331,7 +331,7 @@
'SO2_high ','NH3 ','DMS ','SO2_nat ', &
'BC_biomass','OM_biomass','NO_aircrft']
n_aero_tracers = 12
- n_chem_tracers = 71 ! No chem tracers
+ n_chem_tracers = 73 ! No chem tracers
IF (ukca_config%l_ukca_trophet) THEN
nr_therm = 241 ! thermal reactions
ELSE
@@ -379,7 +379,7 @@
ELSE IF (ukca_config%l_ukca_cristrat) THEN
IF (.NOT. ukca_config%l_ukca_achem) THEN ! Without aerosol
IF (ukca_config%i_ukca_chem_version >= 119) THEN ! CRI-Strat 2 w/o aerosol
- n_chem_tracers = 171 ! advected chemical tracers (+9, -4)
+ n_chem_tracers = 173 ! advected chemical tracers (+9, -4)
n_aero_tracers = 0 ! DMS, SO2... aerosol precursor species
nr_therm = 625 ! thermal reactions (+45 bimol, +8 termol)
nr_phot = 135 ! photolytic reacs
@@ -402,7 +402,7 @@
'MeCO2H ','HOCH2CHO ','NO_aircrft']
ELSE ! With aerosol
IF (ukca_config%i_ukca_chem_version >= 119) THEN ! CRI-Strat 2 with aerosol
- n_chem_tracers = 171 ! advected chemical tracers (+9, -4)
+ n_chem_tracers = 173 ! advected chemical tracers (+9, -4)
n_aero_tracers = 17 ! DMS, SO2... aerosol precursor species
nr_phot = 137 ! photolytic reacs
IF (ukca_config%l_ukca_trophet) THEN
Index: src/science/core/chemistry/ukca_chem_master.F90
===================================================================
--- src/science/core/chemistry/ukca_chem_master.F90 (revision 471)
+++ src/science/core/chemistry/ukca_chem_master.F90 (revision 475)
@@ -103,7 +103,7 @@
! later ones
! Define size of master chemistry
-INTEGER, PARAMETER :: n_chch_master = 356 ! number of known species
+INTEGER, PARAMETER :: n_chch_master = 358 ! number of known species
INTEGER, PARAMETER :: n_het_master = 18 ! number of heterogeneous reactions
INTEGER, PARAMETER :: n_dry_master = 161 ! number of dry deposition reactions
INTEGER, PARAMETER :: n_wet_master = 159 ! number of wet deposition reactions
@@ -917,7 +917,11 @@
! 282
chch_t1(282,'RU10NO3 ',1,'TR ',' ',1,1,cs,0,0,119), &
! 283
-chch_t1(283,'MACO3 ',1,'OO ',' ',0,0,cs,rp,0,119) &
+chch_t1(283,'MACO3 ',1,'OO ',' ',0,0,cs,rp,0,119), &
+! 284
+chch_t1(284,'ALICE ',1,'TR ',' ',0,0,st+cs,0,0,131), &
+! 285
+chch_t1(285,'BOB ',1,'TR ',' ',0,0,st+cs,0,0,131) &
]
Index: src/science/core/chemistry/ukca_constants.F90
===================================================================
--- src/science/core/chemistry/ukca_constants.F90 (revision 471)
+++ src/science/core/chemistry/ukca_constants.F90 (revision 475)
@@ -357,6 +357,10 @@
! Same as Sec_org
REAL, PARAMETER :: c_cri = 5.1782
+! define conversion factors for tutorial species
+REAL, PARAMETER :: c_alice = 1.000
+REAL, PARAMETER :: c_bob = 1.000
+
! molecular masses in g/mol of emitted species,
! for budget calculations
REAL, PARAMETER :: m_ho2 = 33.007
Index: src/science/core/chemistry/ukca_cspecies.F90
===================================================================
--- src/science/core/chemistry/ukca_cspecies.F90 (revision 471)
+++ src/science/core/chemistry/ukca_cspecies.F90 (revision 475)
@@ -182,7 +182,7 @@
c_txcarb22, c_ru12pan, c_rtn26pan, c_aroh14, c_aroh17, c_arnoh14, &
c_arnoh17, c_anhy, c_cri,c_dhpcarb9, c_hpucarb12, c_hucarb9, c_iepox, &
c_hmml, c_dhpr12ooh, c_dhcarb9, c_ru12no3, c_ru10no3, c_dhpr12o2, &
- c_ru10ao2, c_maco3
+ c_ru10ao2, c_maco3, c_alice, c_bob
USE asad_mod, ONLY: advt, nadvt, speci, jpctr, jpspec
@@ -463,6 +463,10 @@
WHERE (advt == 'PASSIVE O3') c_species = 1.0
WHERE (advt == 'AGE OF AIR') c_species = 1.0
+! tutorial species
+WHERE (advt == 'ALICE ') c_species = c_alice
+WHERE (advt == 'BOB ') c_species = c_bob
+
! non-advected tracers
c_na_species=0.0
WHERE (nadvt == 'OH ') c_na_species = c_oh
These differences can be found here:
Tutorials/UMvn13.0/worked_solutions/Task03.1/Task03.1_ukca.patch
Sample output from this task can be found here:
Tutorials/UMvn13.0/sample_output/Task03.1/
Task 3.2: Test your changes in another chemistry scheme
TASK 3.2: Change the chemistry and aero settings to test these changes with StratTrop chemistry, GLOMAP-mode aerosols, and with heterogeneous chemistry back on.
| Solution |
|---|
| Change the value of i_ukca_chem to be 51 (StratTrop) |
| Change the value of l_ukca_chem_aero to be True |
| Ensure that the value of l_ukca_mode is True |
| Change the value of l_ukca_het_psc to be True |
Note that changing these setting will automatically make lots of other changes to variables within Rose. You do not need to worry about these additional changes.
Worked Solution to Task 3.2
The changes to Rose are:
Index: app/ukca/rose-app.conf =================================================================== --- app/ukca/rose-app.conf (revision 236520) +++ app/ukca/rose-app.conf (revision 236523) @@ -477,14 +477,14 @@ !!i_ageair_reset_method=1 !!i_inferno_emi= !!i_mode_bln_param_method=0 -!!i_mode_nzts=15 -!!i_mode_setup=2 -!!i_ukca_activation_scheme=0 -i_ukca_chem=59 +i_mode_nzts=15 +i_mode_setup=2 +i_ukca_activation_scheme=0 +i_ukca_chem=51 i_ukca_chem_version=131 i_ukca_conserve_method=3 -!!i_ukca_dms_flux=1 -!!i_ukca_hetconfig=0 +i_ukca_dms_flux=1 +i_ukca_hetconfig=0 !!i_ukca_hiorder_scheme=0 i_ukca_light_param=1 !!i_ukca_nwbins=20 @@ -507,14 +507,14 @@ !!l_dust_slinn_impc_scav=.false. l_environ_jo2=.false. l_environ_jo2b=.false. -!!l_mode_bhn_on=.true. -!!l_mode_bln_on=.false. +l_mode_bhn_on=.true. +l_mode_bln_on=.false. l_ukca=.true. l_ukca_ageair=.false. !!l_ukca_aie1=.false. !!l_ukca_aie2=.false. l_ukca_asad_columns=.true. -l_ukca_chem_aero=.false. +l_ukca_chem_aero=.true. !!l_ukca_classic_hetchem=.false. !!l_ukca_coarse_no3_prod=.true. l_ukca_ddep_lev1=.false. @@ -522,48 +522,48 @@ l_ukca_debug_asad=.true. l_ukca_dry_dep_so2wet=.false. !!l_ukca_dust=.false. -!!l_ukca_emsdrvn_ch4=.false. +l_ukca_emsdrvn_ch4=.false. !!l_ukca_fine_no3_prod=.true. l_ukca_h2o_feedback=.false. -l_ukca_het_psc=.false. +l_ukca_het_psc=.true. l_ukca_ibvoc=.false. -!!l_ukca_inferno=.false. +l_ukca_inferno=.false. !!l_ukca_inferno_ch4=.false. l_ukca_intdd=.true. l_ukca_intph=.false. -!!l_ukca_limit_nat=.false. +l_ukca_limit_nat=.false. l_ukca_linox_scaling=.false. -!!l_ukca_mode=.false. +l_ukca_mode=.true. l_ukca_prescribech4=.false. !!l_ukca_prim_moc=.false. -!!l_ukca_primbcoc=.false. +l_ukca_primbcoc=.false. !!l_ukca_primdu=.false. -!!l_ukca_primss=.false. -!!l_ukca_primsu=.false. +l_ukca_primss=.false. +l_ukca_primsu=.false. l_ukca_qch4inter=.false. l_ukca_quasinewton=.false. -!!l_ukca_radaer=.false. +l_ukca_radaer=.false. !!l_ukca_radaer_sustrat=.false. l_ukca_radch4=.false. -!!l_ukca_radf11=.false. -!!l_ukca_radf113=.false. -!!l_ukca_radf12=.false. -!!l_ukca_radf22=.false. +l_ukca_radf11=.false. +l_ukca_radf113=.false. +l_ukca_radf12=.false. +l_ukca_radf22=.false. l_ukca_radn2o=.false. l_ukca_rado3=.false. l_ukca_ro2_ntp=.true. l_ukca_ro2_perm=.true. -!!l_ukca_sa_clim=.false. -!!l_ukca_scale_biom_aer_ems=.false. +l_ukca_sa_clim=.false. +l_ukca_scale_biom_aer_ems=.false. !!l_ukca_scale_marine_pom_ems=.false. !!l_ukca_scale_sea_salt_ems=.false. -!!l_ukca_scale_seadms_ems=.false. -!!l_ukca_scale_soa_yield=.true. +l_ukca_scale_seadms_ems=.false. +l_ukca_scale_soa_yield=.true. l_ukca_set_trace_gases=.true. !!l_ukca_sfix=.false. -!!l_ukca_so2ems_expvolc=.false. +l_ukca_so2ems_expvolc=.false. !!l_ukca_src_in_conservation=.true. -!!l_ukca_trophet=.false. +l_ukca_trophet=.false. !!l_ukca_tune_bc=.false. !!l_ukca_use_background_aerosol=.false. lightnox_scale_fac=0.5 @@ -570,10 +570,10 @@ !!marine_pom_ems_scaling=0 !!max_ageair_reset_height=0.0 !!max_ageair_reset_level=0 -!!mode_activation_dryr=37.5 -!!mode_aitsol_cvscav=0.5 -!!mode_incld_so2_rfrac=0.25 -!!mode_parfrac=2.5 +mode_activation_dryr=37.5 +mode_aitsol_cvscav=0.5 +mode_incld_so2_rfrac=0.25 +mode_parfrac=2.5 !!nit=0 nrsteps=45 !!ph_fit_coeff_a=0 @@ -582,7 +582,7 @@ !!phot2d_dir= !!sea_salt_ems_scaling=0 !!seadms_ems_scaling=1.0 -!!soa_yield_scaling=2.0 +soa_yield_scaling=2.0 !!tc_lbc_ukca=150*0 ukca_ccl4mmr=0.00 ukca_cfc115mmr=0.00 @@ -625,9 +625,9 @@ l_fix_ukca_h2dd_x=.false. l_fix_ukca_h2so4_ystore=.true. l_fix_ukca_impscav=.false. -!!l_fix_ukca_meoh_emiss_input=.false. +l_fix_ukca_meoh_emiss_input=.false. !!l_fix_ukca_offox_h2o_fac=.false. -!!l_improve_aero_drydep=.false. +l_improve_aero_drydep=.false. !!l_improve_cv_cons=.false. l_methox_fix=.true. @@ -636,5 +636,5 @@ !!a_convect_segments=-99 !!a_convect_trac_seg_size=1 ukca_chem_seg_size=1 -!!ukca_mode_seg_size=1 +ukca_mode_seg_size=1 ussp_seg_size=1
These differences can be found here:
Tutorials/UMvn13.0/worked_solutions/Task03.2/Task03.2_rose.patch
Sample output from this task can be found here:
Tutorials/UMvn13.0/sample_output/Task03.2/
Checklist
- ☐ Make a ticket at the UKCA trac pages
- ☐ Make a UKCA branch
- ☐ Include this branch within your Rose suite
- ☐ Make your code changes to ukca_chem_master.F90, ukca_config_defs_mod.F90, ukca_constants.F90, and ukca_cspecies.F90
- ☐ Ensure that you are using the correct value of i_ukca_chem_version
- ☐ Test with multiple chemistry schemes as required
UKCA Chemistry and Aerosol Tutorials at UMvn13.0
Written by Luke Abraham 2022