Difference between revisions of "Solution to UKCA Chemistry and Aerosol Tutorial 9 Task 9.1"
(Created page with " Back to UKCA Chemistry and Aerosol Tutorials [[UKCA Chemistry and Aerosol Tutorial 9#Task 9.1: adding new dry deposition values | Back…") |
|||
| (12 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[UKCA Chemistry and Aerosol Tutorials | Back to UKCA Chemistry and Aerosol Tutorials]] |
[[UKCA Chemistry and Aerosol Tutorials | Back to UKCA Chemistry and Aerosol Tutorials]] |
||
| − | [[UKCA Chemistry and Aerosol Tutorial 9#Task 9.1: |
+ | [[UKCA Chemistry and Aerosol Tutorial 9#Task 9.1: Output new diagnostics | Back to the adding new diagnostics tutorial]] |
==Task== |
==Task== |
||
| − | You were asked to |
+ | You were asked to |
<blockquote> |
<blockquote> |
||
| + | Output diagnostics of the reaction <math>\textrm{ALICE} + \textrm{OH} \longrightarrow \textrm{BOB} + \textrm{SEC\_ORG}</math> to STASH code '''34461''', the dry deposition of '''ALICE''' to STASH code '''34462''', and the wet deposition of '''BOB''' to '''34463'''. They should be outputted as a daily mean to the '''pa/UPA''' stream. |
||
| − | You should now add in the dry deposition of '''ALICE'''. This species deposits in a similar way to '''CO'''. The values for '''depvel_defs_strattrop''' are: |
||
| − | |||
| − | {| border="1" |
||
| − | ! Surface Type || Summer (day) || Summer (night) || Summer (24h ave) || Winter (day) || Winter (night) || Winter (24h ave) |
||
| − | |- |
||
| − | | '''Water''' || 0.00 || 0.00 || 0.00 || 0.00 || 0.00 || 0.00 |
||
| − | |- |
||
| − | | '''Forest''' || 0.03 || 0.03 || 0.03 || 0.03 || 0.03 || 0.03 |
||
| − | |- |
||
| − | | '''Grass''' || 0.03 || 0.03 || 0.03 || 0.03 || 0.03 || 0.03 |
||
| − | |- |
||
| − | | '''Desert''' || 0.03 || 0.03 || 0.03 || 0.03 || 0.03 || 0.03 |
||
| − | |- |
||
| − | | '''Ice''' || 0.00 || 0.00 || 0.00 || 0.00 || 0.00 || 0.00 |
||
| − | |} |
||
| − | |||
| − | i.e. the same as for CO. |
||
</blockquote> |
</blockquote> |
||
| Line 30: | Line 14: | ||
<blockquote> |
<blockquote> |
||
| + | Remember to use the correct sampling frequency. |
||
| − | You will need to make changes for both dry deposition schemes. The changes to '''ukca_surfddr.F90''' can be made very easily by adding '''ALICE''' to the '''CO''' block in the <tt>CASE</tt> statement. |
||
</blockquote> |
</blockquote> |
||
==Solution== |
==Solution== |
||
| − | + | As well as making some code changes and making a new user STASHmaster file, you will also need to make some changes to the STASH table to output the diagnostics correctly. |
|
| − | ===Changes to |
+ | ===Changes to asad_flux_dat.F90=== |
| + | The only code changes that need to be made are in '''asad_flux_dat.F90''', where you will need to add the following diagnostic specification blocks |
||
| − | ====chch_defs array==== |
||
| + | asad_flux_defn('RXN',34461,'B',.FALSE.,0,4, & |
||
| − | First, you will need to change the '''0''' in the 6th column of the <tt>chch_defs_strattrop_chem</tt> to '''1''': |
||
| + | (/'ALICE ','OH '/), & |
||
| + | (/'BOB ','Sec_Org ',' ',' '/)), & |
||
| + | asad_flux_defn('DEP',34462,'D',.FALSE.,0,1, & |
||
| + | (/'ALICE ',' '/), & |
||
| + | (/' ',' ',' ',' '/)), & |
||
| + | asad_flux_defn('DEP',34463,'W',.FALSE.,0,1, & |
||
| + | (/'BOB ',' '/), & |
||
| + | (/' ',' ',' ',' '/)) & |
||
| + | It is probably best to make this as a separate array, rather than add this to the end of an existing array. You should then add this array into the '''asad_chemical_fluxes''' master array that holds all the possible diagnostics. You should also increase the '''n_chemical_fluxes''' integer by 3 to '''235''' to take account of these extra fluxes. |
||
| − | chch_t( 76,'ALICE ', 1,'TR ',' ', 1, 0, 0), & ! 76 |
||
| − | === |
+ | ===STASHmaster file=== |
| + | You will need to make a user STASHmaster file which looks similar to |
||
| − | To add in the values for the deposition velocity into the '''depvel_defs_strattrop''' array you will need to |
||
| + | H1| SUBMODEL_NUMBER=1 |
||
| − | * Increase the value of '''ndry_strattrop''' from 36 to '''37''' |
||
| + | H2| SUBMODEL_NAME=ATMOS |
||
| − | * Increase the size of '''depvel_defs_strattrop03''' from 360 to '''390''' |
||
| + | H3| UM_VERSION=8.2 |
||
| − | * Add this code block to end of the '''depvel_defs_strattrop03''' array (remembering to add a comma to end of the line preceding this) |
||
| + | # |
||
| + | #|Model |Sectn | Item |Name | |
||
| + | #|Space |Point | Time | Grid |LevelT|LevelF|LevelL|PseudT|PseudF|PseudL|LevCom| |
||
| + | #| Option Codes | Version Mask | Halo | |
||
| + | #|DataT |DumpP | PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 PC9 PCA | |
||
| + | #|Rotate| PPF | USER | LBVC | BLEV | TLEV |RBLEVV| CFLL | CFFF | |
||
| + | # |
||
| + | #=============================================================================== |
||
| + | # |
||
| + | 1| 1 | 34 | 461 |UKCA Tutorial: ALICE+OH->BOB+Sec_Org| |
||
| + | 2| 0 | 0 | 1 | 1 | 2 | 10 | 11 | 0 | 0 | 0 | 0 | |
||
| + | 3| 000000000000000000000000000000 | 00000000000000000001 | 3 | |
||
| + | 4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | |
||
| + | 5| 0 | 1871 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | |
||
| + | # |
||
| + | 1| 1 | 34 | 462 |UKCA Tutorial: ALICE Dry Dep (3D) | |
||
| + | 2| 0 | 0 | 1 | 1 | 2 | 10 | 11 | 0 | 0 | 0 | 0 | |
||
| + | 3| 000000000000000000000000000000 | 00000000000000000001 | 3 | |
||
| + | 4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | |
||
| + | 5| 0 | 1871 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | |
||
| + | # |
||
| + | 1| 1 | 34 | 463 |UKCA Tutorial: BOB Wet Dep (3D) | |
||
| + | 2| 0 | 0 | 1 | 1 | 2 | 10 | 11 | 0 | 0 | 0 | 0 | |
||
| + | 3| 000000000000000000000000000000 | 00000000000000000001 | 3 | |
||
| + | 4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | |
||
| + | 5| 0 | 1871 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | |
||
| + | # |
||
| + | #=============================================================================== |
||
| + | # |
||
| + | 1| -1 | -1 | -1 |END OF FILE MARK | |
||
| + | 2| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
||
| + | 3| 000000000000000000000000000000 | 00000000000000000000 | 0 | |
||
| + | 4| 0 | 0 | -99 -99 -99 -99 -30 -99 -99 -99 -99 -99 | |
||
| + | 5| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
||
| + | # |
||
| + | This should be included in the UMUI in '''Model Selection → Atmosphere → STASH → User-STASHmaster files. Diags, Progs & Ancils''' and the three diagnostics should be initialised to zero ('''Option''' '''3''') in '''Model Selection → Atmosphere → STASH → Initialisation of User Prognostics'''. |
||
| − | 0.00, 0.00, 0.00, 0.00, 0.00, 0.00, & ! 37.1 |
||
| − | 0.03, 0.03, 0.03, 0.03, 0.03, 0.03, & ! 37.2 |
||
| − | 0.03, 0.03, 0.03, 0.03, 0.03, 0.03, & ! 37.3 |
||
| − | 0.03, 0.03, 0.03, 0.03, 0.03, 0.03, & ! 37.4 |
||
| − | 0.00, 0.00, 0.00, 0.00, 0.00, 0.00 & ! 37.5 |
||
| + | An example STASHmaster file can be found at |
||
| − | ===Changes to ukca_aerod.F90=== |
||
| + | /home/ukca/userprestash/VN8.4/Tutorial/Task9.1_Tutorial_Diags.presm |
||
| − | You will need to add the following block to the '''CASE''' statement within this routine |
||
| + | ===UMUI STASH table changes=== |
||
| − | CASE ('ALICE ') |
||
| − | d0(j) = d_h2o * SQRT(m_h2o / m_ALICE) |
||
| + | Once you have added in your new user STASHmaster file, you can now output these diagnostics through the STASH panel, by placing entries in the STASH table. However, to do this properly you will need to make a new STASH '''Time Profile'''. |
||
| − | the value of m_ALICE having been set in [[Solution to UKCA & UMUI Tutorial 5 Task 5.2#Code changes to ukca_constants.F90 | Task 5.2 - adding new emissions to UKCA]]. |
||
| + | Go to '''Model Selection → Atmosphere → STASH → STASH. Specification of Diagnostics requirements''' to make these changes. |
||
| − | ===Changes to ukca_surfddr.F90=== |
||
| + | ====Create a new time profile==== |
||
| − | As has been noted in the ''hint'' for this task, the only change required to '''ukca_surfddr.F90''' is to add <tt>'ALICE '</tt> to the '''CO''' section of the '''CASE''' statement: |
||
| + | We want to output these diagnostics as daily means, but we cannot use the existing '''TDAYM''' time profile, as the ''sampling frequency'' is incorrect. We also don't want to alter the existing ''TDAYM'' profile as this will then affect all diagnostics currently outputted using this profile. To make the required changes you need to |
||
| − | CASE ('CO ','ALICE ') |
||
| − | rsurf(:,n)=(/3700.,7300.,4550.,1960.,4550.0,r_null,r_null, & |
||
| − | 4550.0,r_null /) ! Shrub+bare soil set to C3 grass (guess) |
||
| − | |||
| − | ===Hand-edit to increase the value of JPDD=== |
||
| + | # Select the '''TDAYM''' profile (it should now highlight yellow) |
||
| − | You will need to add a hand-edit to UMUI in the '''Model Selection → Input/Output Control and Resource → User hand edit files''' by placing it in the table and putting a '''Y''' in the '''Include Y/N''' column. |
||
| + | ## Go to '''Profiles → Copy Profile → Copy time''' |
||
| + | ## You will now be asked for a new name to copy this profile to (e.g. '''TDYMUKCA''') |
||
| + | # Now select this new profile |
||
| + | ## Edit it by going to '''Profiles → Edit Profile → Edit time''' |
||
| + | ## Set the '''Frequency (every)''' to '''3''' |
||
| + | ## Change the '''Sampling offset''' to '''2''' |
||
| + | ## Click '''Close''' to save these changes |
||
| + | You have now created a new time profile with the correct sampling frequency for UKCA diagnostics. |
||
| − | It should contain the following: |
||
| + | ====Output the diagnostics==== |
||
| − | ed CNTLATM<<\EOF |
||
| − | /JPDD/ |
||
| − | d |
||
| − | i |
||
| − | JPDD = 37, |
||
| − | . |
||
| − | w |
||
| − | q |
||
| − | EOF |
||
| + | Now that you have created the new time profile ('''TDYMUKCA'''), you should use it to output the diagnostics as |
||
| − | An example hand-edit can be found at |
||
| + | 34 461 UKCA Tutorial: ALICE+OH->BOB Flux TDYMUKCA DALLTH UPB Y + Y |
||
| − | /home/ukca/hand_edits/VN8.2/Tutorial/Task7.1_incr_JPvals.ed |
||
| + | 34 462 UKCA Tutorial: ALICE Dry Dep (3D) TDYMUKCA DALLTH UPB Y + Y |
||
| + | 34 463 UKCA Tutorial: BOB Wet Dep (3D) TDYMUKCA DALLTH UPB Y + Y |
||
==Output== |
==Output== |
||
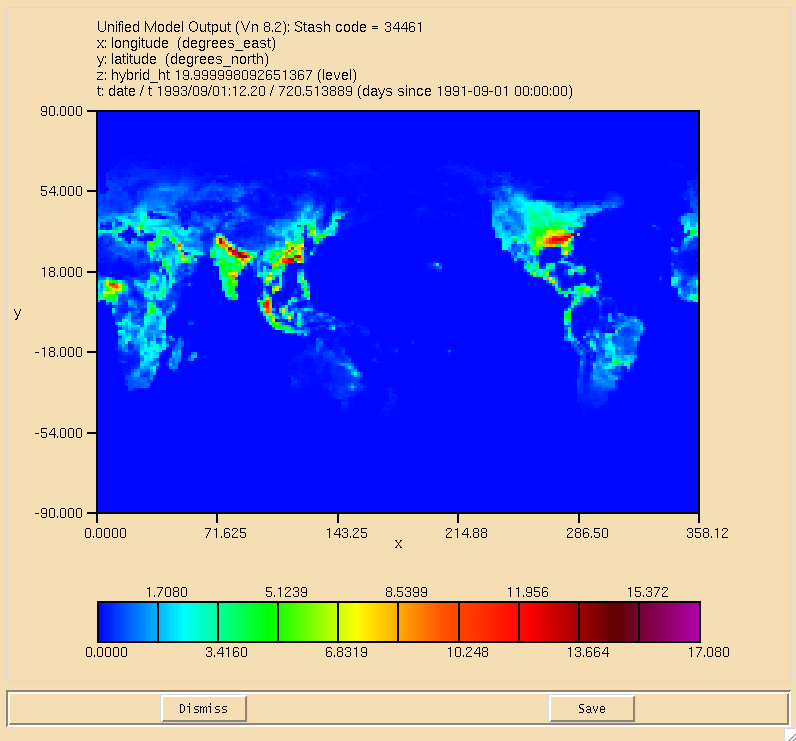
| + | [[Image:ALICE_OH_Flux.png|thumb|150px|right|Figure 1: Flux through the ALICE+OH->BOB Reaction in the lowest model level.]] |
||
| − | If you view the ALICE (34064) field in the '''pb''' file (located in your '''[[UKCA & UMUI Tutorials: Things to know before you start#Archiving|archive]]''' directory) after these changes, and compare it to the equivalent field after the [[Solution to UKCA & UMUI Tutorial 6 Task 6.1 | previous Task (6.1)]] you will see that there are some difference. However, we will not be able to quantify these differences correctly until [[UKCA & UMUI Tutorial 9 | Tutorial 9: Adding new UKCA diagnostics]]. |
||
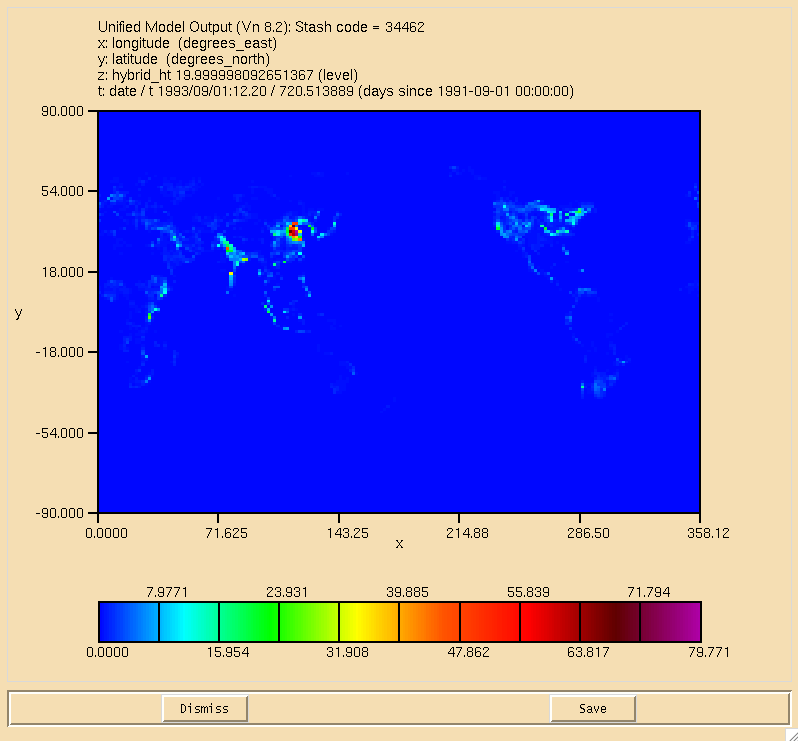
| + | [[Image:ALICE_DryDep.png|thumb|150px|right|Figure 2: Dry deposition of ALICE in the lowest model level.]] |
||
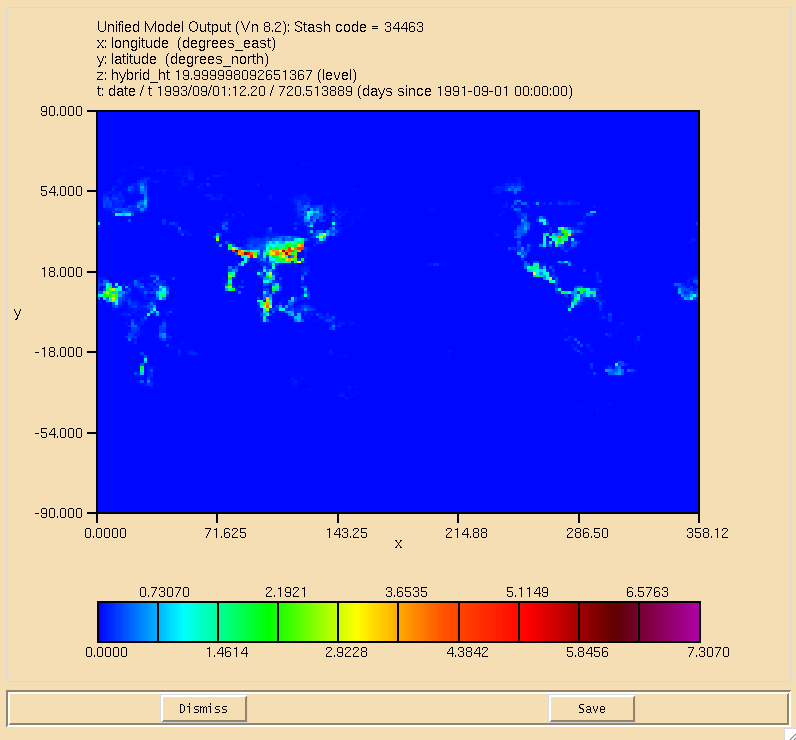
| + | [[Image:BOB_WetDep.png|thumb|150px|right|Figure 3: Wet deposition of BOB in the lowest model level.]] |
||
| + | Your '''pa''' file (located in your job directory) should now contain the following fields |
||
| + | |||
| + | 0 : 192 145 85 1 Stash code = 34001 |
||
| + | 1 : 192 145 85 1 Stash code = 34064 |
||
| + | 2 : 192 145 85 1 Stash code = 34065 |
||
| + | 3 : 192 145 85 1 Stash code = 34461 |
||
| + | 4 : 192 145 85 1 Stash code = 34462 |
||
| + | 5 : 192 145 85 1 Stash code = 34463 |
||
| + | |||
| + | The surface plot of the flux through the <math>\textrm{ALICE} + \textrm{OH} \longrightarrow \textrm{BOB} + \textrm{Sec\_Org}</math> can be seen in Figure 1, the dry deposition flux of ALICE is in Figure 2, and the wet deposition flux of BOB is in Figure 3. All fluxes are in moles/gridcell/s. |
||
'''Sample output''' from this job can be found in |
'''Sample output''' from this job can be found in |
||
| − | /work/n02/n02/ukca/Tutorial/sample_output/ |
+ | /work/n02/n02/ukca/Tutorial/vn8.4/sample_output/Task9.1/ |
| − | on |
+ | on ARCHER, and in |
| − | /projects/ukca/Tutorial/sample_ouput/ |
+ | <strike>/projects/ukca/Tutorial/vn8.4/sample_ouput/Task9.1/</strike> |
on MONSooN. |
on MONSooN. |
||
| Line 112: | Line 143: | ||
==Worked Solution== |
==Worked Solution== |
||
| − | There is a worked solution to this problem in the UMUI Tutorial experiment. This is job ''' |
+ | There is a worked solution to this problem in the UMUI Tutorial experiment. This is job '''j''': ''Tutorial: solution to Task 9.1 - add new diagnostics''. |
| − | The code changes can be viewed by using the following FCM command |
+ | The code changes can be viewed by using the following FCM command |
| − | fcm diff |
+ | fcm diff fcm:um_br/dev/luke/vn8.4_UKCA_Tutorial_Solns@14713 fcm:um_br/dev/luke/vn8.4_UKCA_Tutorial_Solns@14714 |
| − | This gives the following |
+ | This gives the following output: |
| − | Index: src/atmosphere/UKCA/ |
+ | Index: src/atmosphere/UKCA/asad_flux_dat.F90 |
=================================================================== |
=================================================================== |
||
| − | --- src/atmosphere/UKCA/ |
+ | --- src/atmosphere/UKCA/asad_flux_dat.F90 (revision 14713) |
| − | +++ src/atmosphere/UKCA/ |
+ | +++ src/atmosphere/UKCA/asad_flux_dat.F90 (revision 14714) |
| − | @@ - |
+ | @@ -102,7 +102,7 @@ |
| + | CHARACTER(LEN=10) :: blank0 = ' ' ! Defines null product |
||
| − | 500.0, 12500. /) |
||
| − | CASE ('NH3 ') |
||
| − | rsurf(:,n)=hno3 |
||
| − | - CASE ('CO ') |
||
| − | + CASE ('CO ','ALICE ') |
||
| − | rsurf(:,n)=(/3700.,7300.,4550.,1960.,4550.0,r_null,r_null, & |
||
| − | 4550.0,r_null /) ! Shrub+bare soil set to C3 grass (guess) |
||
| − | CASE ('CH4 ') |
||
| − | Index: src/atmosphere/UKCA/ukca_chem_strattrop.F90 |
||
| − | =================================================================== |
||
| − | --- src/atmosphere/UKCA/ukca_chem_strattrop.F90 (revision 12170) |
||
| − | +++ src/atmosphere/UKCA/ukca_chem_strattrop.F90 (revision 12214) |
||
| − | @@ -56,7 +56,7 @@ |
||
| − | INTEGER, PARAMETER, PUBLIC :: nhet_st_tpht = 2 ! trophet rxns |
||
| − | ! |
+ | ! Number of chemical fluxes defined below |
| − | -INTEGER, PARAMETER |
+ | - INTEGER, PARAMETER :: n_chemical_fluxes = 232 |
| − | +INTEGER, PARAMETER |
+ | + INTEGER, PARAMETER :: n_chemical_fluxes = 235 |
| − | INTEGER, PARAMETER, PUBLIC :: ndry_st_aer = 5 ! Aerosol chemistry |
||
| + | ! Do we load standard diagnostics as done below (T), or use only reactions |
||
| − | ! No of wet deposited species |
||
| + | ! specified in STASH (F) ? |
||
| − | @@ -141,7 +141,7 @@ |
||
| + | @@ -923,6 +923,21 @@ |
||
| − | chch_t( 73,'CO2 ', 1,'CT ',' ', 0, 0, 0), & ! 73 |
||
| + | /) |
||
| − | chch_t( 74,'O2 ', 1,'CT ',' ', 0, 0, 0), & ! 74 |
||
| − | chch_t( 75,'N2 ', 1,'CT ',' ', 0, 0, 0), & ! 75 |
||
| − | -chch_t( 76,'ALICE ', 1,'TR ',' ', 0, 0, 0), & ! 76 |
||
| − | +chch_t( 76,'ALICE ', 1,'TR ',' ', 1, 0, 0), & ! 76 |
||
| − | chch_t( 77,'BOB ', 1,'TR ',' ', 0, 0, 0) & ! 77 |
||
| − | /) |
||
| − | @@ -759,7 +759,7 @@ |
||
| + | +! UKCA Tutorial fluxes |
||
| − | REAL, DIMENSION( 360) :: depvel_defs_strattrop01 |
||
| + | + TYPE(asad_flux_defn), PARAMETER, PUBLIC :: & |
||
| − | REAL, DIMENSION( 360) :: depvel_defs_strattrop02 |
||
| + | + UKCA_Tutorial_Fluxes(3) = (/ & |
||
| − | -REAL, DIMENSION( 360) :: depvel_defs_strattrop03 |
||
| + | + asad_flux_defn('RXN',34461,'B',.FALSE.,0,4, & |
||
| − | +REAL, DIMENSION( 390) :: depvel_defs_strattrop03 |
||
| + | + (/'ALICE ','OH '/), & |
||
| − | |||
| + | + (/'BOB ','Sec_Org ',' ',' '/)), & |
||
| − | INTEGER(KIND=jpim), PARAMETER :: zhook_in = 0 |
||
| + | + asad_flux_defn('DEP',34462,'D',.FALSE.,0,1, & |
||
| − | INTEGER(KIND=jpim), PARAMETER :: zhook_out = 1 |
||
| + | + (/'ALICE ',' '/), & |
||
| − | @@ -2120,7 +2120,13 @@ |
||
| − | + | + (/' ',' ',' ',' '/)), & |
|
| − | + | + asad_flux_defn('DEP',34463,'W',.FALSE.,0,1, & |
|
| − | + | + (/'BOB ',' '/), & |
|
| − | + | + (/' ',' ',' ',' '/)) & |
|
| + | + /) |
||
| − | + 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, & ! 36.5 |
||
| + | + |
||
| − | +! 37 ALICE - same as for CO |
||
| + | + |
||
| − | + 0.00, 0.00, 0.00, 0.00, 0.00, 0.00, & ! 37.1 |
||
| + | PUBLIC :: ASAD_LOAD_DEFAULT_FLUXES |
||
| − | + 0.03, 0.03, 0.03, 0.03, 0.03, 0.03, & ! 37.2 |
||
| + | INTERFACE ASAD_LOAD_DEFAULT_FLUXES |
||
| − | + 0.03, 0.03, 0.03, 0.03, 0.03, 0.03, & ! 37.3 |
||
| + | MODULE PROCEDURE ASAD_LOAD_DEFAULT_FLUXES |
||
| − | + 0.03, 0.03, 0.03, 0.03, 0.03, 0.03, & ! 37.4 |
||
| + | @@ -1005,7 +1020,8 @@ |
||
| − | + 0.00, 0.00, 0.00, 0.00, 0.00, 0.00 & ! 37.5 |
||
| + | asad_strat_oh_loss, & ! 26 181 |
||
| − | /) |
||
| + | asad_strat_o3_budget, & ! 20 201 |
||
| − | |||
| + | asad_strat_o3_misc, & ! 15 216 |
||
| − | |||
| + | - asad_aerosol_chem & ! 16 232 |
||
| − | Index: src/atmosphere/UKCA/ukca_aerod.F90 |
||
| + | + asad_aerosol_chem, & ! 16 232 |
||
| − | =================================================================== |
||
| + | + UKCA_Tutorial_Fluxes & ! 3 235 |
||
| − | --- src/atmosphere/UKCA/ukca_aerod.F90 (revision 12170) |
||
| + | /) |
||
| − | +++ src/atmosphere/UKCA/ukca_aerod.F90 (revision 12214) |
||
| + | |||
| − | @@ -222,6 +222,8 @@ |
||
| + | ELSE |
||
| − | d0(j) = d_h2o * SQRT(m_h2o / m_meoh) |
||
| − | CASE ('Monoterp') |
||
| − | d0(j) = d_h2o * SQRT(m_h2o / m_monoterp) |
||
| − | + CASE ('ALICE ') |
||
| − | + d0(j) = d_h2o * SQRT(m_h2o / m_ALICE) |
||
| − | END SELECT |
||
| − | END DO |
||
| − | ! |
||
---- |
---- |
||
Latest revision as of 15:20, 15 December 2015
Back to UKCA Chemistry and Aerosol Tutorials
Back to the adding new diagnostics tutorial
Task
You were asked to
Output diagnostics of the reaction to STASH code 34461, the dry deposition of ALICE to STASH code 34462, and the wet deposition of BOB to 34463. They should be outputted as a daily mean to the pa/UPA stream.
and were given the hint
Remember to use the correct sampling frequency.
Solution
As well as making some code changes and making a new user STASHmaster file, you will also need to make some changes to the STASH table to output the diagnostics correctly.
Changes to asad_flux_dat.F90
The only code changes that need to be made are in asad_flux_dat.F90, where you will need to add the following diagnostic specification blocks
asad_flux_defn('RXN',34461,'B',.FALSE.,0,4, &
(/'ALICE ','OH '/), &
(/'BOB ','Sec_Org ',' ',' '/)), &
asad_flux_defn('DEP',34462,'D',.FALSE.,0,1, &
(/'ALICE ',' '/), &
(/' ',' ',' ',' '/)), &
asad_flux_defn('DEP',34463,'W',.FALSE.,0,1, &
(/'BOB ',' '/), &
(/' ',' ',' ',' '/)) &
It is probably best to make this as a separate array, rather than add this to the end of an existing array. You should then add this array into the asad_chemical_fluxes master array that holds all the possible diagnostics. You should also increase the n_chemical_fluxes integer by 3 to 235 to take account of these extra fluxes.
STASHmaster file
You will need to make a user STASHmaster file which looks similar to
H1| SUBMODEL_NUMBER=1 H2| SUBMODEL_NAME=ATMOS H3| UM_VERSION=8.2 # #|Model |Sectn | Item |Name | #|Space |Point | Time | Grid |LevelT|LevelF|LevelL|PseudT|PseudF|PseudL|LevCom| #| Option Codes | Version Mask | Halo | #|DataT |DumpP | PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 PC9 PCA | #|Rotate| PPF | USER | LBVC | BLEV | TLEV |RBLEVV| CFLL | CFFF | # #=============================================================================== # 1| 1 | 34 | 461 |UKCA Tutorial: ALICE+OH->BOB+Sec_Org| 2| 0 | 0 | 1 | 1 | 2 | 10 | 11 | 0 | 0 | 0 | 0 | 3| 000000000000000000000000000000 | 00000000000000000001 | 3 | 4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | 5| 0 | 1871 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | # 1| 1 | 34 | 462 |UKCA Tutorial: ALICE Dry Dep (3D) | 2| 0 | 0 | 1 | 1 | 2 | 10 | 11 | 0 | 0 | 0 | 0 | 3| 000000000000000000000000000000 | 00000000000000000001 | 3 | 4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | 5| 0 | 1871 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | # 1| 1 | 34 | 463 |UKCA Tutorial: BOB Wet Dep (3D) | 2| 0 | 0 | 1 | 1 | 2 | 10 | 11 | 0 | 0 | 0 | 0 | 3| 000000000000000000000000000000 | 00000000000000000001 | 3 | 4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | 5| 0 | 1871 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | # #=============================================================================== # 1| -1 | -1 | -1 |END OF FILE MARK | 2| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3| 000000000000000000000000000000 | 00000000000000000000 | 0 | 4| 0 | 0 | -99 -99 -99 -99 -30 -99 -99 -99 -99 -99 | 5| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | #
This should be included in the UMUI in Model Selection → Atmosphere → STASH → User-STASHmaster files. Diags, Progs & Ancils and the three diagnostics should be initialised to zero (Option 3) in Model Selection → Atmosphere → STASH → Initialisation of User Prognostics.
An example STASHmaster file can be found at
/home/ukca/userprestash/VN8.4/Tutorial/Task9.1_Tutorial_Diags.presm
UMUI STASH table changes
Once you have added in your new user STASHmaster file, you can now output these diagnostics through the STASH panel, by placing entries in the STASH table. However, to do this properly you will need to make a new STASH Time Profile.
Go to Model Selection → Atmosphere → STASH → STASH. Specification of Diagnostics requirements to make these changes.
Create a new time profile
We want to output these diagnostics as daily means, but we cannot use the existing TDAYM time profile, as the sampling frequency is incorrect. We also don't want to alter the existing TDAYM profile as this will then affect all diagnostics currently outputted using this profile. To make the required changes you need to
- Select the TDAYM profile (it should now highlight yellow)
- Go to Profiles → Copy Profile → Copy time
- You will now be asked for a new name to copy this profile to (e.g. TDYMUKCA)
- Now select this new profile
- Edit it by going to Profiles → Edit Profile → Edit time
- Set the Frequency (every) to 3
- Change the Sampling offset to 2
- Click Close to save these changes
You have now created a new time profile with the correct sampling frequency for UKCA diagnostics.
Output the diagnostics
Now that you have created the new time profile (TDYMUKCA), you should use it to output the diagnostics as
34 461 UKCA Tutorial: ALICE+OH->BOB Flux TDYMUKCA DALLTH UPB Y + Y 34 462 UKCA Tutorial: ALICE Dry Dep (3D) TDYMUKCA DALLTH UPB Y + Y 34 463 UKCA Tutorial: BOB Wet Dep (3D) TDYMUKCA DALLTH UPB Y + Y
Output
Your pa file (located in your job directory) should now contain the following fields
0 : 192 145 85 1 Stash code = 34001 1 : 192 145 85 1 Stash code = 34064 2 : 192 145 85 1 Stash code = 34065 3 : 192 145 85 1 Stash code = 34461 4 : 192 145 85 1 Stash code = 34462 5 : 192 145 85 1 Stash code = 34463
The surface plot of the flux through the can be seen in Figure 1, the dry deposition flux of ALICE is in Figure 2, and the wet deposition flux of BOB is in Figure 3. All fluxes are in moles/gridcell/s.
Sample output from this job can be found in
/work/n02/n02/ukca/Tutorial/vn8.4/sample_output/Task9.1/
on ARCHER, and in
/projects/ukca/Tutorial/vn8.4/sample_ouput/Task9.1/
on MONSooN.
Worked Solution
There is a worked solution to this problem in the UMUI Tutorial experiment. This is job j: Tutorial: solution to Task 9.1 - add new diagnostics.
The code changes can be viewed by using the following FCM command
fcm diff fcm:um_br/dev/luke/vn8.4_UKCA_Tutorial_Solns@14713 fcm:um_br/dev/luke/vn8.4_UKCA_Tutorial_Solns@14714
This gives the following output:
Index: src/atmosphere/UKCA/asad_flux_dat.F90
===================================================================
--- src/atmosphere/UKCA/asad_flux_dat.F90 (revision 14713)
+++ src/atmosphere/UKCA/asad_flux_dat.F90 (revision 14714)
@@ -102,7 +102,7 @@
CHARACTER(LEN=10) :: blank0 = ' ' ! Defines null product
! Number of chemical fluxes defined below
- INTEGER, PARAMETER :: n_chemical_fluxes = 232
+ INTEGER, PARAMETER :: n_chemical_fluxes = 235
! Do we load standard diagnostics as done below (T), or use only reactions
! specified in STASH (F) ?
@@ -923,6 +923,21 @@
/)
+! UKCA Tutorial fluxes
+ TYPE(asad_flux_defn), PARAMETER, PUBLIC :: &
+ UKCA_Tutorial_Fluxes(3) = (/ &
+ asad_flux_defn('RXN',34461,'B',.FALSE.,0,4, &
+ (/'ALICE ','OH '/), &
+ (/'BOB ','Sec_Org ',' ',' '/)), &
+ asad_flux_defn('DEP',34462,'D',.FALSE.,0,1, &
+ (/'ALICE ',' '/), &
+ (/' ',' ',' ',' '/)), &
+ asad_flux_defn('DEP',34463,'W',.FALSE.,0,1, &
+ (/'BOB ',' '/), &
+ (/' ',' ',' ',' '/)) &
+ /)
+
+
PUBLIC :: ASAD_LOAD_DEFAULT_FLUXES
INTERFACE ASAD_LOAD_DEFAULT_FLUXES
MODULE PROCEDURE ASAD_LOAD_DEFAULT_FLUXES
@@ -1005,7 +1020,8 @@
asad_strat_oh_loss, & ! 26 181
asad_strat_o3_budget, & ! 20 201
asad_strat_o3_misc, & ! 15 216
- asad_aerosol_chem & ! 16 232
+ asad_aerosol_chem, & ! 16 232
+ UKCA_Tutorial_Fluxes & ! 3 235
/)
ELSE
Written by Luke Abraham 2014