Difference between revisions of "Solution to UKCA Chemistry and Aerosol Tutorial 5 Task 5.2"
| (8 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[UKCA Chemistry and Aerosol Tutorials | Back to UKCA Chemistry and Aerosol Tutorials]] |
[[UKCA Chemistry and Aerosol Tutorials | Back to UKCA Chemistry and Aerosol Tutorials]] |
||
| − | [[UKCA Chemistry and Aerosol Tutorial 5# |
+ | [[UKCA Chemistry and Aerosol Tutorial 5#Task 5.2: make the required code changes to add your emission into UKCA | Back to the adding new chemical emissions tutorial]] |
==Task== |
==Task== |
||
| Line 8: | Line 8: | ||
<blockquote> |
<blockquote> |
||
| + | You should now make the UKCA code changes to add your emission into the '''ALICE''' tracer. No run-time processing of this surface emission is required. |
||
| − | Make the required code changes so that your '''ALICE''' and '''BOB''' tracers are now specified in the UKCA CheST/StratTrop scheme. You should set the conversion factor for each of these to 1.0. |
||
</blockquote> |
</blockquote> |
||
==Solution== |
==Solution== |
||
| + | As no run-time processing of this field is required, the only changes that need to be made are those in '''ukca_setd1defs.F90''' |
||
| − | The solution to this task involves making changes to 4 files in the UKCA sub-directory of your source code. These files are |
||
| − | + | ===Code changes to ukca_setd1defs.F90=== |
|
| − | ukca_chem_strattrop.F90 |
||
| − | ukca_constants.F90 |
||
| − | ukca_cspecies.F90 |
||
| + | ====em_chem_spec changes==== |
||
| − | The following changes need to be made |
||
| + | In '''ukca_setd1defs.F90''' you should scroll-down to the code block controlled by the |
||
| − | ===Changes to ukca_setd1defs.F90=== |
||
| + | ELSE IF (L_ukca_strattrop .AND. L_ukca_achem) THEN |
||
| − | The value of '''n_chem_tracers''' need to be increased from 71 to '''73'''. |
||
| + | statement and |
||
| − | The '''nm_spec''' array has been edited to add in the ALICE and BOB tracers (changes in <span style="color:red">red</span>): |
||
| + | * '''increase''' the value of '''n_chem_emissions''' to '''22'''. |
||
| − | nm_spec(1:n_all_tracers) = (/ & |
||
| + | * Edit '''em_chem_spec''' so that it now includes '''ALICE''': |
||
| − | 'O3 ','NO ','NO3 ','NO2 ','N2O5 ', & |
||
| − | + | (/'NO ','CH4 ','CO ','HCHO ', & |
|
| − | + | 'C2H6 ','C3H8 ','Me2CO ','MeCHO ', & |
|
| − | + | 'C5H8 ','BC_fossil ','BC_biofuel','OC_fossil ', & |
|
| − | + | 'OC_biofuel','Monoterp ','NVOC ','ALICE ', & |
|
| − | + | 'SO2_low ', & |
|
| − | + | 'SO2_high ','DMS ','NH3 ','SO2_nat ', & |
|
| − | + | 'BC_biomass','OC_biomass','NO_aircrft'/) |
|
| − | 'Cl ','ClO ','Cl2O2 ','OClO ','Br ', & |
||
| − | 'BrO ','BrCl ','BrONO2 ','N2O ','HCl ', & !50 |
||
| − | 'HOCl ','HBr ','HOBr ','ClONO2 ','CFCl3 ', & |
||
| − | 'CF2Cl2 ','MeBr ','N ','O(3P) ','MACRO2 ', & !60 |
||
| − | 'MeCl ','CF2ClBr ','CCl4 ','<span style="color:red">ALICE</span> ','<span style="color:red">BOB</span> ', & |
||
| − | 'MeCCl3 ','CF3Br ','H2OS ','CH2Br2 ','H2 ', & !70 |
||
| − | 'DMS ','SO2 ','H2SO4 ','MSA ','DMSO ', & |
||
| − | 'NH3 ','CS2 ','COS ','H2S ','H ', & !80 |
||
| − | 'OH ','HO2 ','MeOO ','EtOO ','MeCO3 ', & |
||
| − | 'n-PrOO ','i-PrOO ','EtCO3 ','MeCOCH2OO ','MeOH ', & !90 |
||
| − | 'Monoterp ','Sec_Org ','SESQUITERP','SO3 ','AROM ', & |
||
| − | 'O(3P)_S ','O(1D)_S ','NO2 ','BrO ','HCl ', & !100 |
||
| − | 'ND_Nuc_SOL','Nuc_SOL_SU','ND_Ait_SOL','Ait_SOL_SU','Ait_SOL_BC', & |
||
| − | 'Ait_SOL_OC','ND_Acc_SOL','Acc_SOL_SU','Acc_SOL_BC','Acc_SOL_OC', & !110 |
||
| − | 'Acc_SOL_SS','Acc_SOL_DU','ND_Cor_SOL','Cor_SOL_SU','Cor_SOL_BC', & |
||
| − | 'Cor_SOL_OC','Cor_SOL_SS','Cor_SOL_DU','ND_Ait_INS','Ait_INS_BC', & !120 |
||
| − | 'Ait_INS_OC','ND_Acc_INS','Acc_INS_DU','ND_Cor_INS','Cor_INS_Du', & |
||
| − | 'Nuc_SOL_OC','Ait_SOL_SS','Nuc_SOL_OZ','Ait_SOL_OZ','Acc_SOL_OZ', & !130 |
||
| − | 'Cor_SOL_OZ','Nuc_SOL_NH','Ait_SOL_NH','Acc_SOL_NH','Cor_SOL_NH', & |
||
| − | 'Nuc_SOL_NT','Ait_SOL_NT','Acc_SOL_NT','Cor_SOL_NT','XXX ', & !140 |
||
| − | 'Anth_Prec ','Bio_Prec ','Anth_Cond ','Bio_Cond ','XXX ', & |
||
| − | 'XXX ','XXX ','XXX ','PASSIVE O3','AGE OF AIR' & !150 |
||
| − | /) |
||
| + | ====STASH changes==== |
||
| − | ===Changes to ukca_chem_strattrop.F90=== |
||
| + | Edit the IF block controlling which STASH codes are assigned to which species, and add the following |
||
| − | The following lines have been added to the end of the '''chch_defs_strattrop_chem''' specification: |
||
| − | + | ELSEIF (em_chem_spec(i) == 'ALICE ') THEN |
|
| + | UkcaD1Codes(J+i)%item = 316 |
||
| − | chch_t( 77,'BOB ', 1,'TR ',' ', 0, 0, 0) & ! 77 |
||
| + | ===Code changes to ukca_constants.F90=== |
||
| − | (also, a comma has been added after the final ")" on line 75, specifying N2). The size of the <tt>chch_defs_strattrop_chem</tt> array has been increased from 75 to '''77'''. |
||
| + | You need to add the following line to define the molar mass of ALICE |
||
| − | ===Changes to ukca_constants.F90=== |
||
| + | REAL, PARAMETER :: M_ALICE = 28.97 |
||
| − | The following lines have been added to the UKCA_CONSTANTS module: |
||
| + | ===Code changes to ukca_emission_ctl.F90=== |
||
| − | ! UKCA Tutorial tracers |
||
| − | REAL, PARAMETER :: C_ALICE = 1.0000 |
||
| − | REAL, PARAMETER :: C_BOB = 1.0000 |
||
| + | You need to add the following line to the WHERE block adding in the molar masses of the emitted species (defined in ''em_chem_spec'') to the '''molmass''' array |
||
| − | ===Changes to ukca_cspecies.F90=== |
||
| + | WHERE (em_chem_spec == 'ALICE ') molmass = M_ALICE |
||
| − | The following lines have been added to the UKCA_CALC_CSPECIES subroutine which is held within the UKCA_CSPECIES module |
||
| − | |||
| − | ! UKCA Tutorial Tracers |
||
| − | WHERE (advt == 'ALICE ') c_species = C_ALICE |
||
| − | WHERE (advt == 'BOB ') c_species = C_BOB |
||
| − | |||
| − | ===Hand-edit to increase values of JPCTR and JPSPEC=== |
||
| − | |||
| − | A hand-edit is required to increase the values of '''JPCTR''' and '''JPSPEC'''. This should contain the following: |
||
| − | |||
| − | ed CNTLATM<<\EOF |
||
| − | /JPCTR/ |
||
| − | d |
||
| − | i |
||
| − | JPCTR = 73, |
||
| − | . |
||
| − | /JPSPEC/ |
||
| − | d |
||
| − | i |
||
| − | JPSPEC = 77, |
||
| − | . |
||
| − | w |
||
| − | q |
||
| − | EOF |
||
| − | |||
| − | An example can be found at |
||
| − | |||
| − | /home/ukca/hand_edits/VN8.2/Tutorial/Task4.2_incr_JPvals.ed |
||
==Output== |
==Output== |
||
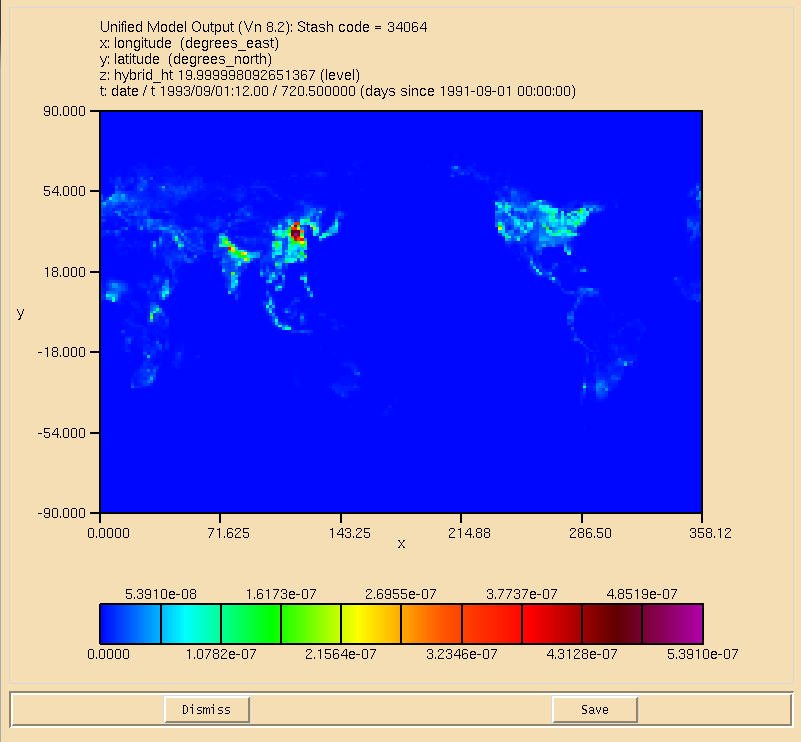
| + | [[Image:UKCA_Tutorial_ALICE_Surface.png|thumb|300px|right|Figure 1: Surface plot of the ALICE tracer after emissions have been applied.]] |
||
| − | If you open the '''pb''' file in your '''[[UKCA & UMUI Tutorials: Things to know before you start#Archiving|archive]]''' directory you will find that it still contains the fields |
||
| + | As the ALICE (and BOB) tracers are being output as daily means to the pb/UPB stream, you can examine the output (located in your '''[[UKCA & UMUI Tutorials: Things to know before you start#Archiving|archive]]''' directory) to see if the emission is being correctly applied. Opening the '''pa''' file there are 3 fields: |
||
0 : 192 145 85 1 Stash code = 34001 |
0 : 192 145 85 1 Stash code = 34001 |
||
| Line 117: | Line 63: | ||
2 : 192 145 85 1 Stash code = 34065 |
2 : 192 145 85 1 Stash code = 34065 |
||
| − | + | The first (34001) is ozone, and 34065 is BOB (which will still contain zeros). View the surface of 34064 and you should now see that it is non-zero. An example of this is shown in Figure 1. |
|
| + | |||
| + | You should note that this will not match up exactly with the emission field in the ancillary file, partly because of the time interpolation that is done between the monthly emission fields, and partly because the field in the tracer is a daily mean of a tracer that has these emissions applied every timestep (and currently with no loss processes), and which is also undergoing boundary layer mixing to mix concentrations out of the surface layer. |
||
'''Sample output''' from this job can be found in |
'''Sample output''' from this job can be found in |
||
| − | /work/n02/n02/ukca/Tutorial/sample_output/ |
+ | /work/n02/n02/ukca/Tutorial/vn8.4/sample_output/Task5.2/ |
| − | on |
+ | on ARCHER, and in |
| − | /projects/ukca/Tutorial/sample_ouput/ |
+ | <strike>/projects/ukca/Tutorial/vn8.4/sample_ouput/Task5.2/</strike> |
on MONSooN. |
on MONSooN. |
||
| Line 131: | Line 79: | ||
==Worked Solution== |
==Worked Solution== |
||
| − | There is a worked solution to this problem in the UMUI Tutorial experiment. This is job ''' |
+ | There is a worked solution to this problem in the UMUI Tutorial experiment. This is job '''f''': ''Tutorial: solution to Task 5.2 - adding new chemical emissions in UKCA''. |
The code changes can be viewed by using the following FCM command |
The code changes can be viewed by using the following FCM command |
||
| − | fcm diff |
+ | fcm diff fcm:um_br/dev/luke/vn8.4_UKCA_Tutorial_Solns@14692 fcm:um_br/dev/luke/vn8.4_UKCA_Tutorial_Solns@14697 |
This gives the following (non-graphical) output: |
This gives the following (non-graphical) output: |
||
| Line 141: | Line 89: | ||
Index: src/atmosphere/UKCA/ukca_setd1defs.F90 |
Index: src/atmosphere/UKCA/ukca_setd1defs.F90 |
||
=================================================================== |
=================================================================== |
||
| − | --- src/atmosphere/UKCA/ukca_setd1defs.F90 (revision |
+ | --- src/atmosphere/UKCA/ukca_setd1defs.F90 (revision 14692) |
| − | +++ src/atmosphere/UKCA/ukca_setd1defs.F90 (revision |
+ | +++ src/atmosphere/UKCA/ukca_setd1defs.F90 (revision 14697) |
| − | @@ - |
+ | @@ -273,14 +273,15 @@ |
| + | nr_phot = 55 ! photolytic (ATA) |
||
| + | |||
| + | ELSE IF (L_ukca_strattrop .AND. L_ukca_achem) THEN |
||
| + | - n_chem_emissions = 21 ! em_chem_spec below |
||
| + | + n_chem_emissions = 22 ! em_chem_spec below |
||
| + | n_3d_emissions = 2 ! volc SO2 & aircraft NOX |
||
| + | ALLOCATE(em_chem_spec(n_chem_emissions+n_3d_emissions)) |
||
| + | em_chem_spec = & |
||
(/'NO ','CH4 ','CO ','HCHO ', & |
(/'NO ','CH4 ','CO ','HCHO ', & |
||
'C2H6 ','C3H8 ','Me2CO ','MeCHO ', & |
'C2H6 ','C3H8 ','Me2CO ','MeCHO ', & |
||
| − | 'C5H8 ',' |
+ | 'C5H8 ','BC_fossil ','BC_biofuel','OC_fossil ', & |
| − | - |
+ | - 'OC_biofuel','Monoterp ','NVOC ','SO2_low ', & |
| − | + |
+ | + 'OC_biofuel','Monoterp ','NVOC ','ALICE ', & |
| − | + | + 'SO2_low ', & |
|
| − | + | 'SO2_high ','DMS ','NH3 ','SO2_nat ', & |
|
| + | 'BC_biomass','OC_biomass','NO_aircrft'/) |
||
| − | |||
| + | n_aero_tracers = 12 |
||
| − | @@ -409,7 +409,7 @@ |
||
| + | @@ -624,6 +625,8 @@ |
||
| − | 'BrO ','BrCl ','BrONO2 ','N2O ','HCl ', & !50 |
||
| + | UkcaD1Codes(J+i)%item = 314 |
||
| − | 'HOCl ','HBr ','HOBr ','ClONO2 ','CFCl3 ', & |
||
| − | + | ELSEIF (em_chem_spec(i) == 'NVOC ') THEN |
|
| + | UkcaD1Codes(J+i)%item = 315 |
||
| − | - 'MeCl ','CF2ClBr ','CCl4 ','CF2ClCFCl2','CHF2Cl ', & |
||
| − | + |
+ | + ELSEIF (em_chem_spec(i) == 'ALICE ') THEN |
| + | + UkcaD1Codes(J+i)%item = 316 |
||
| − | 'MeCCl3 ','CF3Br ','H2OS ','CH2Br2 ','H2 ', & !70 |
||
| + | ELSEIF (em_chem_spec(i) == 'BC_biomass') THEN |
||
| − | 'DMS ','SO2 ','H2SO4 ','MSA ','DMSO ', & |
||
| + | UkcaD1Codes(J+i)%item = 322 |
||
| − | 'NH3 ','CS2 ','COS ','H2S ','H ', & !80 |
||
| + | UkcaD1Codes(J+i)%len_dim3 = tr_levels |
||
Index: src/atmosphere/UKCA/ukca_constants.F90 |
Index: src/atmosphere/UKCA/ukca_constants.F90 |
||
=================================================================== |
=================================================================== |
||
| − | --- src/atmosphere/UKCA/ukca_constants.F90 (revision |
+ | --- src/atmosphere/UKCA/ukca_constants.F90 (revision 14692) |
| − | +++ src/atmosphere/UKCA/ukca_constants.F90 (revision |
+ | +++ src/atmosphere/UKCA/ukca_constants.F90 (revision 14697) |
| − | @@ - |
+ | @@ -385,6 +385,11 @@ |
| − | REAL, PARAMETER :: |
+ | REAL, PARAMETER :: m_alkaooh = 90.0 |
| − | REAL, PARAMETER :: |
+ | REAL, PARAMETER :: m_aromooh = 130.0 |
| + | REAL, PARAMETER :: m_mekooh = 104.0 |
||
| − | |||
| − | +! UKCA Tutorial tracers |
||
| − | + REAL, PARAMETER :: C_ALICE = 1.0000 |
||
| − | + REAL, PARAMETER :: C_BOB = 1.0000 |
||
| − | + |
||
+ |
+ |
||
| + | +! UKCA Tutorial ALICE tracer - same as mass of air in g/mol |
||
| − | ! molecular masses in g/mol of emitted species, |
||
| + | +! as the value of C_ALICE=1.0000 (above) |
||
| − | ! for budget calculations |
||
| + | +! Required for ukca_emission_ctl |
||
| − | |||
| + | + REAL, PARAMETER :: M_ALICE = 28.97 |
||
| − | Index: src/atmosphere/UKCA/ukca_chem_strattrop.F90 |
||
| + | |||
| + | ! The mass of organic nitrate is an approximation, |
||
| + | ! calculated as the average of ORGNIT formed by two |
||
| + | Index: src/atmosphere/UKCA/ukca_emission_ctl.F90 |
||
=================================================================== |
=================================================================== |
||
| − | --- src/atmosphere/UKCA/ |
+ | --- src/atmosphere/UKCA/ukca_emission_ctl.F90 (revision 14692) |
| − | +++ src/atmosphere/UKCA/ |
+ | +++ src/atmosphere/UKCA/ukca_emission_ctl.F90 (revision 14697) |
| − | @@ - |
+ | @@ -380,6 +380,8 @@ |
| + | WHERE (em_chem_spec == 'OC_fossil ') molmass = m_c |
||
| + | WHERE (em_chem_spec == 'OC_biofuel') molmass = m_c |
||
| + | WHERE (em_chem_spec == 'OC_biomass') molmass = m_c |
||
| + | +! UKCA Tutorial Tracer |
||
| + | + WHERE (em_chem_spec == 'ALICE ') molmass = M_ALICE |
||
| + | ! Check if all the emitted species have a valid molecular weight |
||
| − | |||
| + | IF (ANY(molmass(:) < 0.00001)) THEN |
||
| − | ! ATA NLA CheST Chemistry v1.2 |
||
| − | -TYPE(CHCH_T), DIMENSION( 75), PUBLIC :: chch_defs_strattrop_chem=(/ & |
||
| − | +TYPE(CHCH_T), DIMENSION( 77), PUBLIC :: chch_defs_strattrop_chem=(/ & |
||
| − | chch_t( 1,'O(3P) ', 1,'TR ','Ox ', 0, 0, 0), & ! 1 |
||
| − | chch_t( 2,'O(1D) ', 1,'SS ','Ox ', 0, 0, 0), & ! 2 |
||
| − | chch_t( 3,'O3 ', 1,'TR ','Ox ', 1, 0, 0), & ! 3 DD: 1, |
||
| − | @@ -140,7 +140,9 @@ |
||
| − | chch_t( 72,'MeOH ', 1,'TR ',' ', 1, 1, 0), & ! 72 DD:36,WD:29, |
||
| − | chch_t( 73,'CO2 ', 1,'CT ',' ', 0, 0, 0), & ! 73 |
||
| − | chch_t( 74,'O2 ', 1,'CT ',' ', 0, 0, 0), & ! 74 |
||
| − | -chch_t( 75,'N2 ', 1,'CT ',' ', 0, 0, 0) & ! 75 |
||
| − | +chch_t( 75,'N2 ', 1,'CT ',' ', 0, 0, 0), & ! 75 |
||
| − | +chch_t( 76,'ALICE ', 1,'TR ',' ', 0, 0, 0), & ! 76 |
||
| − | +chch_t( 77,'BOB ', 1,'TR ',' ', 0, 0, 0) & ! 77 |
||
| − | /) |
||
| − | |||
| − | TYPE(CHCH_T), DIMENSION( 87), PUBLIC :: chch_defs_strattrop_aer=(/ & |
||
| − | Index: src/atmosphere/UKCA/ukca_cspecies.F90 |
||
| − | =================================================================== |
||
| − | --- src/atmosphere/UKCA/ukca_cspecies.F90 (revision 12143) |
||
| − | +++ src/atmosphere/UKCA/ukca_cspecies.F90 (revision 12148) |
||
| − | @@ -270,6 +270,9 @@ |
||
| − | WHERE (advt == 'ORGNIT ') c_species = c_orgnit |
||
| − | WHERE (advt == 'PASSIVE O3') c_species = 1.0 |
||
| − | WHERE (advt == 'AGE OF AIR') c_species = 1.0 |
||
| − | +! UKCA Tutorial Tracers |
||
| − | + WHERE (advt == 'ALICE ') c_species = C_ALICE |
||
| − | + WHERE (advt == 'BOB ') c_species = C_BOB |
||
| − | |||
| − | ! non-advected tracers |
||
| − | c_na_species=0.0 |
||
| − | |||
Latest revision as of 15:11, 15 December 2015
Back to UKCA Chemistry and Aerosol Tutorials
Back to the adding new chemical emissions tutorial
Task
You were asked to
You should now make the UKCA code changes to add your emission into the ALICE tracer. No run-time processing of this surface emission is required.
Solution
As no run-time processing of this field is required, the only changes that need to be made are those in ukca_setd1defs.F90
Code changes to ukca_setd1defs.F90
em_chem_spec changes
In ukca_setd1defs.F90 you should scroll-down to the code block controlled by the
ELSE IF (L_ukca_strattrop .AND. L_ukca_achem) THEN
statement and
- increase the value of n_chem_emissions to 22.
- Edit em_chem_spec so that it now includes ALICE:
(/'NO ','CH4 ','CO ','HCHO ', &
'C2H6 ','C3H8 ','Me2CO ','MeCHO ', &
'C5H8 ','BC_fossil ','BC_biofuel','OC_fossil ', &
'OC_biofuel','Monoterp ','NVOC ','ALICE ', &
'SO2_low ', &
'SO2_high ','DMS ','NH3 ','SO2_nat ', &
'BC_biomass','OC_biomass','NO_aircrft'/)
STASH changes
Edit the IF block controlling which STASH codes are assigned to which species, and add the following
ELSEIF (em_chem_spec(i) == 'ALICE ') THEN
UkcaD1Codes(J+i)%item = 316
Code changes to ukca_constants.F90
You need to add the following line to define the molar mass of ALICE
REAL, PARAMETER :: M_ALICE = 28.97
Code changes to ukca_emission_ctl.F90
You need to add the following line to the WHERE block adding in the molar masses of the emitted species (defined in em_chem_spec) to the molmass array
WHERE (em_chem_spec == 'ALICE ') molmass = M_ALICE
Output
As the ALICE (and BOB) tracers are being output as daily means to the pb/UPB stream, you can examine the output (located in your archive directory) to see if the emission is being correctly applied. Opening the pa file there are 3 fields:
0 : 192 145 85 1 Stash code = 34001 1 : 192 145 85 1 Stash code = 34064 2 : 192 145 85 1 Stash code = 34065
The first (34001) is ozone, and 34065 is BOB (which will still contain zeros). View the surface of 34064 and you should now see that it is non-zero. An example of this is shown in Figure 1.
You should note that this will not match up exactly with the emission field in the ancillary file, partly because of the time interpolation that is done between the monthly emission fields, and partly because the field in the tracer is a daily mean of a tracer that has these emissions applied every timestep (and currently with no loss processes), and which is also undergoing boundary layer mixing to mix concentrations out of the surface layer.
Sample output from this job can be found in
/work/n02/n02/ukca/Tutorial/vn8.4/sample_output/Task5.2/
on ARCHER, and in
/projects/ukca/Tutorial/vn8.4/sample_ouput/Task5.2/
on MONSooN.
Worked Solution
There is a worked solution to this problem in the UMUI Tutorial experiment. This is job f: Tutorial: solution to Task 5.2 - adding new chemical emissions in UKCA.
The code changes can be viewed by using the following FCM command
fcm diff fcm:um_br/dev/luke/vn8.4_UKCA_Tutorial_Solns@14692 fcm:um_br/dev/luke/vn8.4_UKCA_Tutorial_Solns@14697
This gives the following (non-graphical) output:
Index: src/atmosphere/UKCA/ukca_setd1defs.F90
===================================================================
--- src/atmosphere/UKCA/ukca_setd1defs.F90 (revision 14692)
+++ src/atmosphere/UKCA/ukca_setd1defs.F90 (revision 14697)
@@ -273,14 +273,15 @@
nr_phot = 55 ! photolytic (ATA)
ELSE IF (L_ukca_strattrop .AND. L_ukca_achem) THEN
- n_chem_emissions = 21 ! em_chem_spec below
+ n_chem_emissions = 22 ! em_chem_spec below
n_3d_emissions = 2 ! volc SO2 & aircraft NOX
ALLOCATE(em_chem_spec(n_chem_emissions+n_3d_emissions))
em_chem_spec = &
(/'NO ','CH4 ','CO ','HCHO ', &
'C2H6 ','C3H8 ','Me2CO ','MeCHO ', &
'C5H8 ','BC_fossil ','BC_biofuel','OC_fossil ', &
- 'OC_biofuel','Monoterp ','NVOC ','SO2_low ', &
+ 'OC_biofuel','Monoterp ','NVOC ','ALICE ', &
+ 'SO2_low ', &
'SO2_high ','DMS ','NH3 ','SO2_nat ', &
'BC_biomass','OC_biomass','NO_aircrft'/)
n_aero_tracers = 12
@@ -624,6 +625,8 @@
UkcaD1Codes(J+i)%item = 314
ELSEIF (em_chem_spec(i) == 'NVOC ') THEN
UkcaD1Codes(J+i)%item = 315
+ ELSEIF (em_chem_spec(i) == 'ALICE ') THEN
+ UkcaD1Codes(J+i)%item = 316
ELSEIF (em_chem_spec(i) == 'BC_biomass') THEN
UkcaD1Codes(J+i)%item = 322
UkcaD1Codes(J+i)%len_dim3 = tr_levels
Index: src/atmosphere/UKCA/ukca_constants.F90
===================================================================
--- src/atmosphere/UKCA/ukca_constants.F90 (revision 14692)
+++ src/atmosphere/UKCA/ukca_constants.F90 (revision 14697)
@@ -385,6 +385,11 @@
REAL, PARAMETER :: m_alkaooh = 90.0
REAL, PARAMETER :: m_aromooh = 130.0
REAL, PARAMETER :: m_mekooh = 104.0
+
+! UKCA Tutorial ALICE tracer - same as mass of air in g/mol
+! as the value of C_ALICE=1.0000 (above)
+! Required for ukca_emission_ctl
+ REAL, PARAMETER :: M_ALICE = 28.97
! The mass of organic nitrate is an approximation,
! calculated as the average of ORGNIT formed by two
Index: src/atmosphere/UKCA/ukca_emission_ctl.F90
===================================================================
--- src/atmosphere/UKCA/ukca_emission_ctl.F90 (revision 14692)
+++ src/atmosphere/UKCA/ukca_emission_ctl.F90 (revision 14697)
@@ -380,6 +380,8 @@
WHERE (em_chem_spec == 'OC_fossil ') molmass = m_c
WHERE (em_chem_spec == 'OC_biofuel') molmass = m_c
WHERE (em_chem_spec == 'OC_biomass') molmass = m_c
+! UKCA Tutorial Tracer
+ WHERE (em_chem_spec == 'ALICE ') molmass = M_ALICE
! Check if all the emitted species have a valid molecular weight
IF (ANY(molmass(:) < 0.00001)) THEN
Written by Luke Abraham 2014