UKCA Chemistry and Aerosol Tutorial 1
Back to UKCA Chemistry and Aerosol Tutorials
What you will do in this tutorial
In this tutorial you will now take a copy of a UKCA job and send it to compile and run on the supercomputer. We will then learn about how to check if the job is running, and also about some of the output files produced.
Getting hold of an existing UM-UKCA job
Before you can start using UKCA (and the UM generally), you first need to copy a UM or UKCA job from somewhere. You may be given a job from someone for a specific purpose, or you may just copy a standard job. In the UMUI there are two repositories of standard jobs, under the umui and ukca UMUI users. The umui user contains a series of standard jobs provided and maintained by NCAS-CMS. The ukca user contains jobs provided by the UKCA team.
Running an existing UKCA job
You will need to change a number of options within the UMUI to allow you to run this job successfully, such as your username, ARCHER TIC-code (if needed) etc. If you are using the MONSooN job you may also need to change the project group in
Model Selection -> Post Processing -> Main Switch + General Questions
if you want to send output data to the /nerc data disk (this is advisable). The NCAS-CMS UMUI Training Video will give you the minimum information that you need to be able to make these changes.
Task 1.1: Copy a UM-UKCA job and then run it
More details on how to copy UMUI jobs can be found in the NCAS-CMS Introduction to the UMUI tutorial video.
TASK 1.1: Make a new experiment and take a copy of the UKCA Tutorial Base Job, filter for the ukca user and choose the correct experiment for the machine that you will be running on (ARCHER: xjrn, MONSooN: xjrj). Select the a job, labeled Tutorial: Base UM-UKCA Chemistry and Aerosol Job and copy this one to your own experiment.
Now take your copy of the Tutorial Base Job and make the required changes to allow this job to run. Once you have made these changes you can submit your job. First click Save, then Process, and once this has completed, click Submit. This will then extract the code from the FCM repositories and submit them to the supercomputer. If you are running on MONSooN you will need to enter your passcode at this stage.
Note: To allow the jobs in this tutorial to run quickly this job is only set to run for 2 days. This means that there will be no climate-mean files produced (see the what is STASH? tutorial) produced, which require run lengths of a month or more.
Sample output from this job can be found in
/work/n02/n02/ukca/Tutorial/vn8.4/sample_output/Base/
on ARCHER, and in
/projects/ukca/Tutorial/vn8.4/sample_ouput/Base/
on MONSooN.
Checking the progress of a running job
Log-in to the supercomputer, and check that your job is running. For ARCHER do
qstat -u $USER
and for MONSooN do
llq -u $USER
This should give a list of your running jobs. For example, on ARCHER you get output similar to
$ qstat -u $USER
sdb:
Req'd Req'd Elap
Job ID Username Queue Jobname SessID NDS TSK Memory Time S Time
--------------- -------- -------- ---------- ------ --- --- ------ ----- - -----
1515659.sdb luke par:8n_2 xjqka_run 7934 1 1 -- 00:10 R 00:05
and on MONSooN you should get something like
$ llq -u $USER Id Owner Submitted ST PRI Class Running On ------------------------ ---------- ----------- -- --- ------------ ----------- mon001.64641.0 nlabra 6/5 12:36 R 50 parallel c139 1 job step(s) in query, 0 waiting, 0 pending, 1 running, 0 held, 0 preempted
You can also check how far a job has gone while it is running. To do this you will need to cd into the job directory (this will be on your /work space on ARCHER or your /projects space on MONSooN). When you do this, you will see something like this
$ ls baserepos/ history_archive/ umatmos/ umscripts/ xjlla.list xjlla.stash xjlla.xhist xjllaa.pa20051202 bin/ pe_output/ umrecon/ xjlla.astart xjlla.requests xjlla.umui.nl xjllaa.pa20051201
Now cd into the pe_output/ directory and do
$ tail -f jobid.fort6.pe0 | grep Atm_Step
Atm_Step: Timestep 67 Model time: 2005-12-01 22:20:00
Atm_Step: Timestep 68 Model time: 2005-12-01 22:40:00
Atm_Step: Timestep 69 Model time: 2005-12-01 23:00:00
Atm_Step: Timestep 70 Model time: 2005-12-01 23:20:00
Atm_Step: Timestep 71 Model time: 2005-12-01 23:40:00
Atm_Step: Timestep 72 Model time: 2005-12-02 00:00:00
(changing jobid as appropriate for your job).
Viewing and extracting output
To take a look at the output, you will need to change into your job directory. Once in this directory ls to see the file listing. All the output data is contained in files with the naming convention of
jobida.pzYYYYMMDD
(e.g. xjllaa.pa20051201) or
jobida.pzYYYYmmm
(e.g. xjcina.pb2006dec). The what is STASH? tutorial will discuss the various output streams (denoted by the letter z above) will be discussed in more detail.
Restart files have a similar naming strategy:
jobida.daYYYYMMDD_HH
(e.g. xjcina.da20070201_00). These files are known as dumps.
To see what output files have been produced, do
$ ls *.p* xjllaa.pa20051201
As you can see, there is one file present, the "pa" file. This file is a daily file that has come from the UPA PP stream (standard PP files will be covered in more detail in the What is STASH? tutorial. To quickly view output you can use Xconv, which provides a simple data viewer. It can also be used to convert the UM format output files to netCDF.
You can open these files by
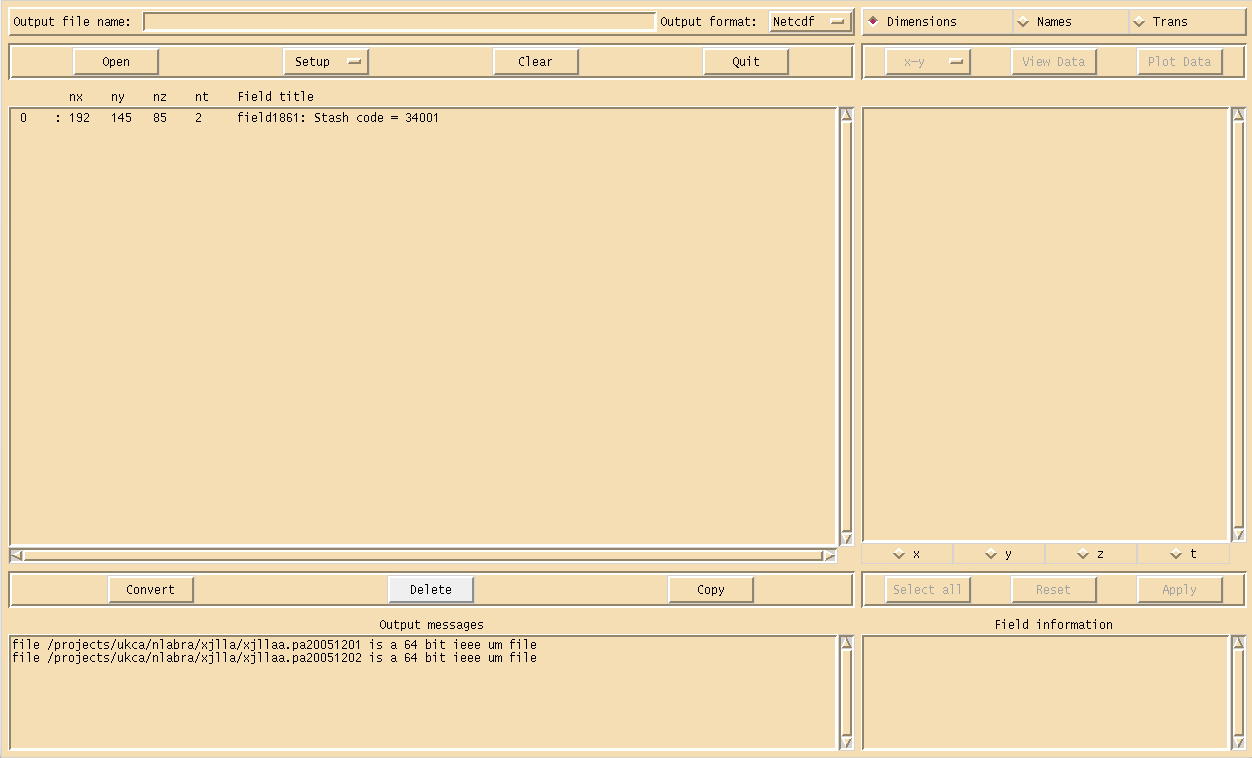
$ xconv -i *.pa*
which will show the Xconv window as can been seen in Figure 1. There is only one field present
0 : 192 145 85 1 Stash code = 34001
This is the UKCA chemical ozone tracer (although it is not labeled as such by default). A full listing of all UKCA fields can be found in the listing of UKCA fields at UM8.2 (which is the same in UM8.4). More information will be given on STASH in the What is STASH? tutorial.
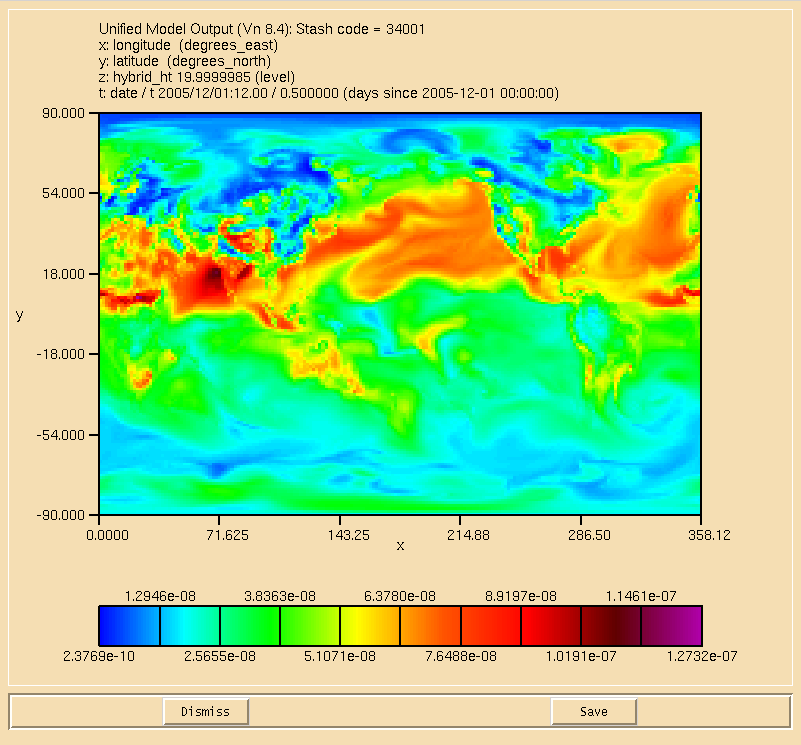
You can use Xconv to view certain fields. For example, you could view the surface ozone concentration double-clicking on the Stash code = 34001 field and clicking the Plot data button (see Figure 2). While this is good to quickly check data, the plotting functions are rather limited as it is not possible to change e.g. the colour-bar, the scale, add a map projection etc. It is advisable to either export fields as netCDF from within Xconv, or to use another program, such as IDL (using the Met Office library) or Python (using either cf-python or Iris) which is able to read the UM PP/FieldsFile format directly.
To export fields as netCDF select them using the mouse (they should then highlight blue), enter a name for the netCDF file in the Output file name box (making sure that the Output format is Netcdf) and click the Convert button. The window on the bottom right will show the progress of the conversion. For single fields this is usually quite quick, but it is possible to use Xconv to open multiple files containing a series of times. In this case Xconv will combine all the individual times into a single field, and outputting this can take some time.
One issue you may have is that Xconv uses a quantity called the field code to determine the variable name of each field (the netCDF name attribute). For UKCA tracer fields at UM8.2 this code is all the same, so all variables will be called field1861. It is possible to change the short field name in Xconv, prior to outputting a netCDF file. Select the variable you wish to output and select the Names button on the top-right of the Xconv window. Delete the contents of the short field name box and replace it with what you would like, e.g. for ozone (Stash code 34001) you may wish to use the CF standard name mass_fraction_of_ozone_in_air (as the units of UKCA tracers are kg(species)/kg(air)). The click apply and output the field as normal. When running ncdump on the resultant netCDF file you should see something like
float mass_fraction_of_ozone_in_air(t, hybrid_ht, latitude, longitude) ;
mass_fraction_of_ozone_in_air:source = "Unified Model Output (Vn 8.4):" ;
mass_fraction_of_ozone_in_air:name = "mass_fraction_of_ozone_in_air" ;
mass_fraction_of_ozone_in_air:title = "Stash code = 34001" ;
mass_fraction_of_ozone_in_air:date = "01/12/05" ;
mass_fraction_of_ozone_in_air:time = "00:00" ;
mass_fraction_of_ozone_in_air:long_name = "Stash code = 34001" ;
mass_fraction_of_ozone_in_air:units = " " ;
mass_fraction_of_ozone_in_air:missing_value = 2.e+20f ;
mass_fraction_of_ozone_in_air:_FillValue = 2.e+20f ;
mass_fraction_of_ozone_in_air:valid_min = 2.29885e-10f ;
mass_fraction_of_ozone_in_air:valid_max = 1.839324e-05f ;
Once you have your data as netCDF it is then possible to use any standard visualisation or processing package to view and manipulate the data.
ncdump
On MONSooN ncdump is located in
/home/accowa/MTOOLS/bin
and on ARCHER to use ncdump you will first need to
module swap PrgEnv-cray PrgEnv-gnu module load nco
and it should then be available in your $PATH. You should not put these lines in your .profile or .bashrc file however, as this may affect your use of the UM.
.leave Files
The text output from any write statements within the code, or giving information about compilation, is outputted to several files with the extension .leave. These will be in your $HOME/output directory on ARCHER and MONSooN.
You will have three .leave files, one for the compilation, one for the reconfiguration step (if run), and one for the UM itself. By default for climate runs these will all have a common format, starting with 4 blocks of letters and numbers, like this:
xipfa000.xipfa.d13163.t120017
where this breaks down to
| jobidXXX | e.g. xipfa000 | The jobid of the job, followed by the job-step number. For compilation and reconfiguration jobs, this will be 000, but as the CRUN progresses this number will increment by 1 for each step, and then cycle round back through 000 (if you run more than 999 steps). |
| jobid | e.g. xipfa | The jobid of the job as listed in the UMUI. |
| dXXXXX | e.g. 13163 | The year (the last two digits, i.e. 2013 is 13) and the day of the year as 3 digits (i.e. 001-366, so this file was created on the 12th June (day 163)). |
| tXXXXXX | e.g. 120017 | The time in HHMMSS format, as recorded by the system clock on the supercomputer. |
Using this format this means that file was created on the 12th June 2013 at 12:00:17. Note that the timestamp on the file will be later than this, as this is the time the file was created, not the time that it was last written to.
There are then three file extensions: .comp.leave for compilation output, .rcf.leave for reconfiguration output, and .leave for the model output.
It is often easier to list your files in this directory by date, but using ls -ltr.
Compilation Output (.comp.leave)
This gives the output from either the XLF compiler on MONSooN or the Cray compiler on ARCHER. If the compilation step has an error and the code is not compiled you can find the source of the error by opening this file and searching for failed - this will highlight which routine(s) caused the problem. You may also get more detailed information such as the line number which had the error. In this case you can open the file on the supercomputer and view the line, as the line number given will not match with the line in your working directory on PUMA due to merging source code and the use of include files. Remember to make any required changes to your PUMA source code however!
Reconfiguration Output (.rcf.leave)
This gives output from the reconfiguration step, if run. At older UM versions, such as UM7.3 this output was found in the model output .leave file.
Model Output (.leave)
This gives output from the code which is generated as it is running, although this file is only updated and closed when the job finishes. To view the output generated as it is running you will need to see the output in the pe_output/ directory mentioned above.
To run efficiently the UM is split into many domains, which communicate with each other with parallel calls, during runtime. The exact decomposition is defined in Model Selection → User Information and Submit Method → Job submission method, in the number of processes East-West and North South boxes. If you have a 12x8 decomposition there will be 96 processes, running on 96 cores of the supercomputer (3 nodes of MONSooN, 4 nodes of ARCHER). These processes will be numbered internally from 0 → 95, labelled as PE0 to e.g. PE95. Only the output from PE0 will be sent to the .leave file, with output from the other PEs only held in the pe_output/ directory. Whether or not these files are deleted at the end of a run is set in the UMUI in Model Selection → Input/Output Control and Resources → Output management panel. If you run fails then these files will not be deleted.
While there is a lot of information outputted to the .leave file, and you would usually only read it if the job fails, it is worth going through the messages, making a special note of any warnings.
If you see the line
jobid: Run terminated normally
around line 48 then your job has successfully run.
Written by Luke Abraham 2014