UKCA Chemistry and Aerosol vn11.8 Tutorial 5
UKCA Chemistry and Aerosol Tutorials at vn11.8
What you will learn in this Tutorial
During this tutorial you will learn how to make new UKCA netCDF emissions files. Then you will learn how to add new emissions into UKCA so that they emit into one of your new tracers.
At the end of the previous tutorial you will now know how to create new tracers for use by UKCA. However, after completing the tasks, your tracers will still be empty, as nothing has been put into them. This tutorial will teach you how to create an emissions file that the UKCA will read, and that you can then tell UKCA to use and emit into your tracer(s).
This tutorial will go through the steps needed to make an emission into a tracer which UKCA does not currently emit into. The steps in making the netCDF file will be the same for a species which is currently emitted into, although in this simpler case you would not need to make any code changes.
This example only deals with a single field. For a more detailed examples on changing emissions for all species, please see the Emissions for ACSIS webpage.
Task 5.1: Create a new emissions file and use it in your job
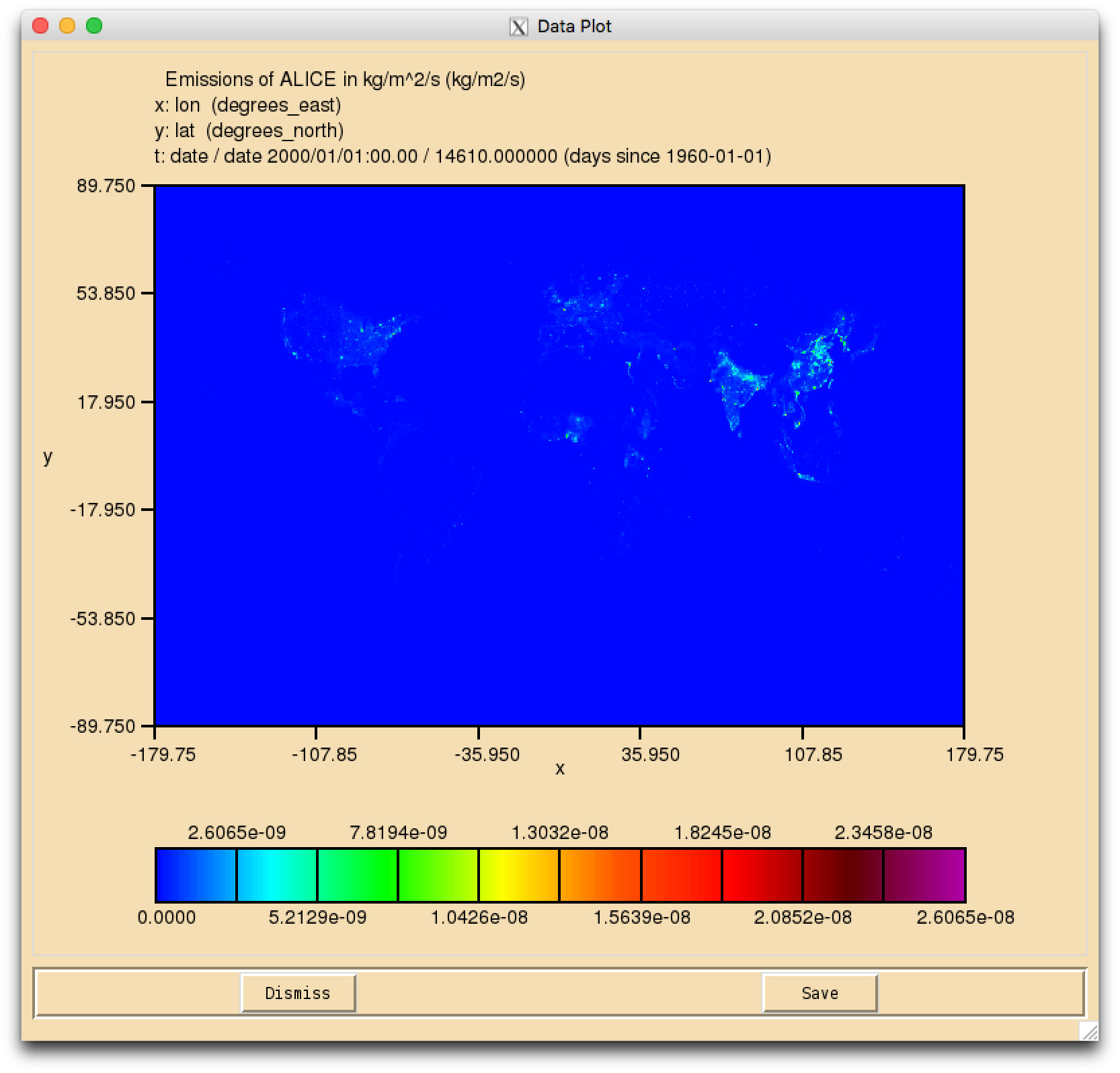
Task 5.1: In theTutorials/vn11.8/Task05.1directory there is the file Emissions_of_ALICE.nc which is a 0.5x0.5 degree resolution surface emission field. You should regrid this file to the N48 ENDGame grid, and output it as a netCDF file that has the required netCDF metadata for a climatological surface emission without any diurnal cycle.
The netCDF emissions system
UKCA uses netCDF files to specify the emissions into UKCA species. Using netCDF files means that the metadata in the file can be used to specify various options, such as which diurnal cycle to use.
Metadata
In Global Atmosphere 7.0 and above, UKCA emissions are prescribed using NetCDF files, rather than using the the UM ancillary file format as NetCDF files allow for more flexibility and include informative metadata.
Each emission field in the NetCDF files needs to include the following variable metadata attributes (there is no requirement for the netCDF variable names themselves except that they be no longer than 80 characters):
standard_name: It should be included if an appropriate name is present in the CF Standard Name Table (see http://cfconventions.org/standard-names.html). An example of a valid ‘standard name’ is“tendency_of_atmosphere_mass_content_of_nitrogen_monoxide_due_to_emission”
(note that all substrings have to be separated by underscores).long_name: It is recommendable but only compulsory if there is no ‘standard_name’ available for that emission field. There are no specific CF conventions for standard names, but within the UKCA code all substrings should be separated by white space as shown in this example:“tendency of atmosphere mass content of nitrogen monoxide due to emission”.
The reason for this is given in the description of the attribute units.tracer_name: This attribute has to be equal to the name of a tracer (or of an emission field such as ‘NO - aircrft’) present in the list of emissions for the given chemical scheme, i.e.em_chem_spec. This name is used by the subroutine UKCA_ADD_EMISS to add the emission field to the corresponding tracer. There should be at least one emission field in the NetCDF files (or alternatively an online emission field) with the value of tracer name equal to one of the items in em_chem_spec, otherwise the subroutine UKCA_EMISS_INIT (in module UKCA_EMISS_MOD) will report a missing tracer in the emission files and the model will stop with error.units: According to CF conventions, all emissions should be expressed in . As a consequence, all files will contain the attributeunits = "kg m-2 s-1"
However, under some circumstances emission fields need to be reported as kg of a given species (e.g. nitrogen, carbon, sulfur). When that is the case this needs to be indicated in the standard name attribute if possible (see example below), otherwise in the long name attribute. As an example, in the case of an emission field reported as kg of carbon, the attribute units will be “kg m-2 s-1” while the attribute standard name should contain the substring“expressed_as_carbon”if that is accepted by CF conventions for standard names (see http://cf-pcmdi.llnl.gov/documents/cf-standard-names); otherwise the attribute long name should contain the substring“expressed as carbon”. The UM subroutines BASE_EMISS_FACTORS and GET_BASE_SCALING will look for such substrings and apply some conversions if needed. It is therefore essential that the substrings in standard_name and long_name are separated by underscores and white spaces, respectively, as indicated above.hourly_scaling: (optional) A character attribute used to apply a diurnal cycle to emissions data with daily or lower frequency. Allowed values:none(or attribute not present): no scalingtraffic_uk: used for UK air quality studiesTNO_MACC_EU_SNAPnn(where nn=01 to 11): Hourly factors of emissions for Europe. Calculated by TNO for the MACC project.diurnal_isopems(for isoprene emissions): use the routine UKCA DIURNAL ISOP EMS to calculate a diurnal cycle using solar zenith angle and latitude to compute the expected number of sunshine hours.
daily_scaling(optional): A character attribute used to apply a weekly cycle to emissions weekly or lower frequency. Only allowed when the model is using the Gregorian calendar. Allowed values:none(or attribute not present): no scalingtraffic_uk: used for UK air quality studiesTNO_MACC_GB_SNAPnn(where nn=01 to 11): Hourly factors of emissions for Great Britain. Calculated by TNO for the MACC project.
vertical_scaling(optional): A character attribute defining the vertical distribution of the emission. This allows the user to supply a 2D field to be applied over multiple levels. Allowed values:surface(or attribute not present): treat as surface emissionall_levels,3D: field is three-dimensional and is provided on model levels (must have the same number of levels as the model)high_level: spread a 2D field over multiple model levels, weighting by model layer thickness to achieve a uniform distribution in height. Must be accompanied by variable attributes lowest level and highest level to indicate the model levels over which to distribute the emission.step1: used for air quality simulations, spreads emission over lowest 3 layers of the model; only allowed with 38 model levels.EMEP_modified_SNAPnn(where nn=01 to 11): Average vertical profiles for SNAP sectors, similar to implementation for EMEP model. See routine vertical emiss factors for details.Bieser_modified_SNAPnn(where nn=01 to 11): Average vertical profiles for SNAP sectors, similar to implementation of Bieser et al. (2011) with the SMOKE model and including fugitive emissions. See routine vertical_emiss_factors for details. In addition, each NetCDF file has to include two global attributes with information that is valid for all emission fields present in the file:
update_freq_in_hours: Integer indicating the frequency (in hours) at which all emission fields present in that file should be read to update emissions(:)%values (:,:,:) in the UKCA code. Update points are calculated relative the model’s ancil reftime.update_type: Integer number indicating the times at which the data is provided. The same conventions as for ancillary files have been adopted:- 0: Single time
- 1: Time series
- 2: Periodic time series
- When emissions are treated as time series (update_type=1) the user is responsible for creating emission fields which cover the whole period for which the model is run; otherwise the model will not find the time registers to do interpolations and will stop with error. When emissions are periodic (update_type=2) each emission field has to include exactly 12 monthly average emissions (Jan, Feb, ..., Dec). Other attributes, in particular some global attributes, as well as some additional fields (e.g. a variable indicating the type of grid mapping) should be present in the files to comply with CF conventions for NetCDF data, but they are not used by the UKCA code.
Using python to regrid your emissions
For this task we will be using the Iris python library, which is developed by the Met Office and can read Met Office formatted files. The cf-python library also does this, but we will not be using this here.
If you have been provided a VM, Iris will have been installed for you, and can be accessed by using the command
pyterm
The command
pylab
has also been provided to give you an iPython environment which logs the commands you type and has included the --pylab flag.
Personal Virtual Machine
To be able to use Iris on the VM, you should first edit the script
/usr/local/bin/install-iris
to use the command
conda install -y -c conda-forge python=3.6 iris=1.13 ipython mo_pack numpy=1.15
which installs the required python packages from anaconda, and also creates an alias in your ~/.bashrc file, called conda, which opens a terminal with additional paths set. You may need to re-source your ~/.bashrc by
. ~/.bashrc
before running the
conda
command.
Using python
when developing python scripts, a handy command to use can be set by the following alias:
alias pylab='ipython --pylab --logfile=ipython-`date +"%Y%m%d-%H%M%S"`.py'
This uses ipython, which is an interactive shell for python. The --pylab automatically loads some standard python libraries such as scipy, numpy, and matplotlib. The --logfile means that all commands will be recorded in a file with the format ipython-YYYYMMDD-HHMMSS.py. If you have been provided a VM, this command has already be defined for you.
You can also execute pre-written python scripts by
- vm: launch iPython (e.g. via
pytermfollowed bypylab) and then run the script using
%run "/path/to/script.py"
However, don't panic. You are not expected to know python to complete this tutorial. You will be provided with an example script to use that you only need to edit a few lines to get to work. You do not need to write a script from scratch, just read-through the provided script and try to understand what it does, and why.
Example python script
In the directory:
Tutorials/vn11.8/Task05.1
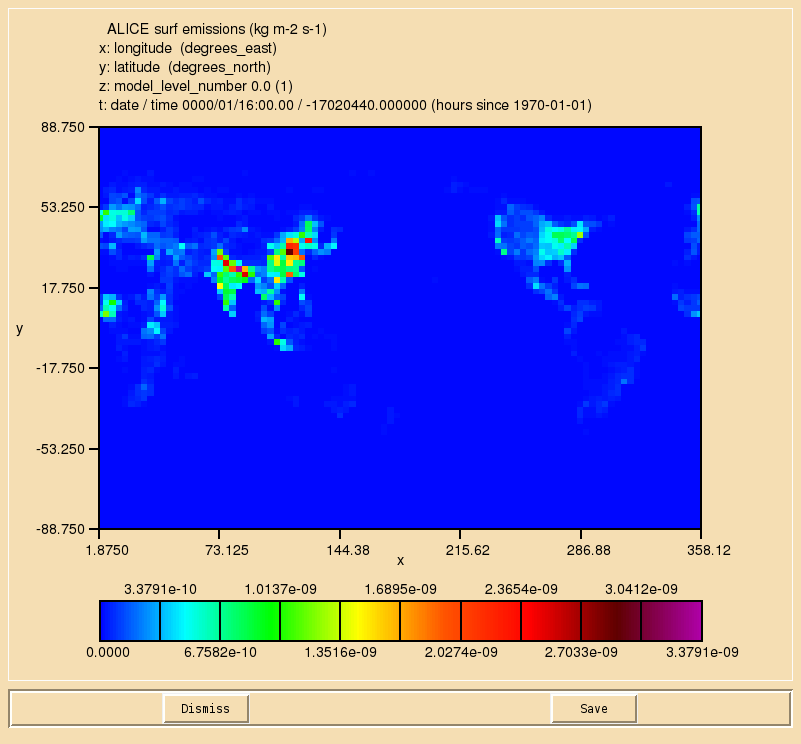
you will find the file regrid_ALICE_N48e.py. You should take a copy of this file, and using your output .pa file from your suite, you should regrid the emissions into the N48 ENDGame grid. You will later use the resulting netCDF file in your suite.
This file looks like:
#!/usr/bin/env python # This file is part of the UKCA Tutorials: # http://www.ukca.ac.uk/wiki/index.php/UKCA_Chemistry_and_Aerosol_Tutorials_at_vn11.8 # Copyright (C) 2021 University of Cambridge # This is free software: you can redistribute it and/or modify it under the # terms of the GNU Lesser General Public License as published by the Free Software # Foundation, either version 3 of the License, or (at your option) any later # version. # It is distributed in the hope that it will be useful, but WITHOUT ANY # WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A # PARTICULAR PURPOSE. See the GNU Lesser General Public License for more details. # You find a copy of the GNU Lesser General Public License at <http://www.gnu.org/licenses/>. # Written by N. Luke Abraham 2021-01-04 <nla27@cam.ac.uk> # To use this script on a personal VM you should first # install-iris # and then open a new terminal with the correct PATHs set using the command # conda # If you have been provided a VM, please use the command # pyterm # followed by # pylab # once you have done this, you can run the script (through ipython) using # %run "/path/to/regrid_ALICE_N48e.py" # preamble import iris import numpy # pre-Iris v1.10, use iris.unit instead of cf_units import cf_units # --- CHANGE THINGS BELOW THIS LINE TO WORK WITH YOUR FILES ETC. --- # name of file containing an ENDGame grid, e.g. your model output # NOTE: all the fields in the file should be on the same horizontal # grid, as the field used MAY NOT be the first in order of STASH grid_file='/umshared/Tutorials/vn11.8/sample_output/Base/atmosa.pa19810901_00' # name of emissions file emissions_file='/umshared/Tutorials/vn11.8/Task05.1/Emissions_of_ALICE.nc' # --- BELOW THIS LINE, NOTHING SHOULD NEED TO BE CHANGED --- species_name='ALICE' # this is the grid we want to regrid to, e.g. N48 ENDGame grd=iris.load_cube(grid_file,iris.AttributeConstraint(STASH='m01s34i001')) grd.coord(axis='x').guess_bounds() grd.coord(axis='y').guess_bounds() # This is the original data ems=iris.load_cube(emissions_file) # make intersection between 0 and 360 longitude to ensure that # the data is regridded correctly nems = ems.intersection(longitude=(0, 360)) # make sure that we use the same coordinate system, otherwise regrid won't work nems.coord(axis='x').coord_system=grd.coord_system() nems.coord(axis='y').coord_system=grd.coord_system() # now guess the bounds of the new grid prior to regridding nems.coord(axis='x').guess_bounds() nems.coord(axis='y').guess_bounds() # now regrid ocube=nems.regrid(grd,iris.analysis.AreaWeighted()) # now add correct attributes and names to netCDF file ocube.var_name='emissions_'+str.strip(species_name) ocube.long_name=str.strip(species_name)+' surf emissions' ocube.units=cf_units.Unit('kg m-2 s-1') ocube.attributes['vertical_scaling']='surface' ocube.attributes['tracer_name']=str.strip(species_name) # global attributes, so don't set in local_keys # NOTE: all these should be strings, including the numbers! This will change at a later UM version. # basic emissions type ocube.attributes['emission_type']='2' # periodic ocube.attributes['update_type']='2' # same as above ocube.attributes['update_freq_in_hours']='120' # i.e. 5 days ocube.attributes['source']='UKCA Tutorial Task 5.1 - creating netCDF emissions' # rename and set time coord - set to be 0000/01/16:00:00-0000/12/16:00:00 # this bit is annoyingly fiddly ocube.coord(axis='t').var_name='time' ocube.coord(axis='t').standard_name='time' ocube.coords(axis='t')[0].units=cf_units.Unit('hours since 1970-01-01', calendar='360_day') ocube.coord(axis='t').points=numpy.array([-17020440, -17019720, -17019000, -17018280, -17017560, -17016840, -17016120, -17015400, -17014680, -17013960, -17013240, -17012520]) # make z-direction. zdims=iris.coords.DimCoord(numpy.array([0]),standard_name = 'model_level_number', units='1',attributes={'positive':'up'}) ocube.add_aux_coord(zdims) ocube=iris.util.new_axis(ocube, zdims) # now transpose cube to put Z 2nd ocube.transpose([1,0,2,3]) # make coordinates 64-bit ocube.coord(axis='x').points=ocube.coord(axis='x').points.astype(dtype='float64') ocube.coord(axis='y').points=ocube.coord(axis='y').points.astype(dtype='float64') #ocube.coord(axis='z').points=ocube.coord(axis='z').points.astype(dtype='float64') # integer ocube.coord(axis='t').points=ocube.coord(axis='t').points.astype(dtype='float64') # for some reason, longitude_bounds are double, but latitude_bounds are float ocube.coord('latitude').bounds=ocube.coord('latitude').bounds.astype(dtype='float64') # add forecast_period & forecast_reference_time # forecast_reference_time frt=numpy.array([-17020080, -17019360, -17018640, -17017920, -17017200, -17016480, -17015760, -17015040, -17014320, -17013600, -17012880, -17012160],dtype='float64') frt_dims=iris.coords.AuxCoord(frt,standard_name = 'forecast_reference_time', units=cf_units.Unit('hours since 1970-01-01', calendar='360_day')) ocube.add_aux_coord(frt_dims,data_dims=0) ocube.coord('forecast_reference_time').guess_bounds() # forecast_period fp=numpy.array([-360],dtype='float64') fp_dims=iris.coords.AuxCoord(fp,standard_name = 'forecast_period', units=cf_units.Unit('hours'),bounds=numpy.array([-720,0],dtype='float64')) ocube.add_aux_coord(fp_dims,data_dims=None) # add-in cell_methods ocube.cell_methods = [iris.coords.CellMethod('mean', 'time')] # set _FillValue fillval=1e+20 ocube.data = numpy.ma.array(data=ocube.data, fill_value=fillval, dtype='float32') # output file name, based on species outpath='ukca_emiss_'+species_name+'.nc' # don't want time to be cattable, as is a periodic emissions file iris.FUTURE.netcdf_no_unlimited=True # annoying hack to set a missing_value attribute as well as a _FillValue attribute dict.__setitem__(ocube.attributes, 'missing_value', fillval) # now write-out to netCDF saver = iris.fileformats.netcdf.Saver(filename=outpath, netcdf_format='NETCDF3_CLASSIC') saver.update_global_attributes(Conventions=iris.fileformats.netcdf.CF_CONVENTIONS_VERSION) saver.write(ocube, local_keys=['vertical_scaling', 'missing_value','um_stash_source','tracer_name']) # end of script # Why we are messing around with metadata? #----------------------------------------- # # We need to adapt the metadata of the emissions data to # match what UKCA is expecting. # e.g. the metadata of the 'Emissions_of_ALICE.nc file is: # # netcdf Emissions_of_ALICE { # dimensions: # lon = 720 ; # lat = 360 ; # date = UNLIMITED ; // (12 currently) # variables: # float lon(lon) ; # lon:long_name = "Longitude" ; # lon:standard_name = "longitude" ; # lon:units = "degrees_east" ; # lon:point_spacing = "even" ; # lon:modulo = " " ; # float lat(lat) ; # lat:long_name = "Latitude" ; # lat:standard_name = "latitude" ; # lat:units = "degrees_north" ; # lat:point_spacing = "even" ; # float date(date) ; # date:long_name = "Time" ; # date:units = "days since 1960-01-01" ; # date:time_origin = "01-JAN-1960:00:00:00" ; # float ALICE(date, lat, lon) ; # ALICE:source = " " ; # ALICE:name = "ALICE" ; # ALICE:title = "Emissions of ALICE in kg/m^2/s" ; # ALICE:date = "01/01/60" ; # ALICE:time = "00:00" ; # ALICE:long_name = "Emissions of ALICE in kg/m^2/s" ; # ALICE:standard_name = "tendency_of_atmosphere_mass_content_of_ALICE_due_to_emission" ; # ALICE:units = "kg/m2/s" ; # ALICE:missing_value = 2.e+20f ; # ALICE:_FillValue = 2.e+20f ; # ALICE:valid_min = 0.f ; # ALICE:valid_max = 2.60646e-08f ; # # // global attributes: # :history = "Tue Jun 18 14:32:42 BST 2013 - XCONV V1.92 16-February-2006" ; # } # # whereas, the metadata of the # /home/vagrant/umdir/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_CO.nc # file is, e.g.: # # netcdf ukca_emiss_CO { # dimensions: # time = UNLIMITED ; // (12 currently) # model_level_number = 1 ; # latitude = 72 ; # longitude = 96 ; # bnds = 2 ; # variables: # double emissions_CO(time, model_level_number, latitude, longitude) ; # emissions_CO:long_name = "CO surf emissions" ; # emissions_CO:units = "kg m-2 s-1" ; # emissions_CO:um_stash_source = "m01s00i303" ; # emissions_CO:tracer_name = "CO" ; # emissions_CO:vertical_scaling = "surface" ; # emissions_CO:cell_methods = "time: mean" ; # emissions_CO:grid_mapping = "latitude_longitude" ; # emissions_CO:coordinates = "forecast_period forecast_reference_time" ; # int latitude_longitude ; # latitude_longitude:grid_mapping_name = "latitude_longitude" ; # latitude_longitude:longitude_of_prime_meridian = 0. ; # latitude_longitude:earth_radius = 6371229. ; # double time(time) ; # time:axis = "T" ; # time:bounds = "time_bnds" ; # time:units = "hours since 1970-01-01 00:00:00" ; # time:standard_name = "time" ; # time:calendar = "360_day" ; # double time_bnds(time, bnds) ; # int model_level_number(model_level_number) ; # model_level_number:axis = "Z" ; # model_level_number:units = "metre" ; # model_level_number:standard_name = "model_level_number" ; # model_level_number:long_name = "height at theta layer midpoint" ; # model_level_number:positive = "up" ; # float latitude(latitude) ; # latitude:axis = "Y" ; # latitude:bounds = "latitude_bnds" ; # latitude:units = "degrees_north" ; # latitude:standard_name = "latitude" ; # float latitude_bnds(latitude, bnds) ; # float longitude(longitude) ; # longitude:axis = "X" ; # longitude:bounds = "longitude_bnds" ; # longitude:units = "degrees_east" ; # longitude:standard_name = "longitude" ; # double longitude_bnds(longitude, bnds) ; # double forecast_period ; # forecast_period:bounds = "forecast_period_bnds" ; # forecast_period:units = "hours" ; # forecast_period:standard_name = "forecast_period" ; # double forecast_period_bnds(bnds) ; # double forecast_reference_time(time) ; # forecast_reference_time:units = "hours since 1970-01-01 00:00:00" ; # forecast_reference_time:standard_name = "forecast_reference_time" ; # forecast_reference_time:calendar = "360_day" ; # # // global attributes: # :emission_type = "2" ; # :source = "Data from Met Office Unified Model" ; # :um_version = "7.3" ; # :update_freq_in_hours = "120" ; # :update_type = "2" ; # :Conventions = "CF-1.5" ; # } # # so the metadata of our new emissions file needs to be edited to be what UKCA # expects. # # After using this script, the resultant netCDF file should look like: # # netcdf ukca_emiss_ALICE { # dimensions: # time = 12 ; # model_level_number = 1 ; # latitude = 72 ; # longitude = 96 ; # bnds = 2 ; # variables: # float emissions_ALICE(time, model_level_number, latitude, longitude) ; # emissions_ALICE:_FillValue = 1.e+20f ; # emissions_ALICE:long_name = "ALICE surf emissions" ; # emissions_ALICE:units = "kg m-2 s-1" ; # emissions_ALICE:missing_value = 1.e+20 ; # emissions_ALICE:tracer_name = "ALICE" ; # emissions_ALICE:vertical_scaling = "surface" ; # emissions_ALICE:cell_methods = "time: mean" ; # emissions_ALICE:grid_mapping = "latitude_longitude" ; # emissions_ALICE:coordinates = "forecast_period forecast_reference_time" ; # int latitude_longitude ; # latitude_longitude:grid_mapping_name = "latitude_longitude" ; # latitude_longitude:longitude_of_prime_meridian = 0. ; # latitude_longitude:earth_radius = 6371229. ; # double time(time) ; # time:axis = "T" ; # time:units = "hours since 1970-01-01" ; # time:standard_name = "time" ; # time:long_name = "Time" ; # time:calendar = "360_day" ; # time:time_origin = "01-JAN-1960:00:00:00" ; # int model_level_number(model_level_number) ; # model_level_number:axis = "Z" ; # model_level_number:units = "1" ; # model_level_number:standard_name = "model_level_number" ; # model_level_number:positive = "up" ; # double latitude(latitude) ; # latitude:axis = "Y" ; # latitude:bounds = "latitude_bnds" ; # latitude:units = "degrees_north" ; # latitude:standard_name = "latitude" ; # double latitude_bnds(latitude, bnds) ; # double longitude(longitude) ; # longitude:axis = "X" ; # longitude:bounds = "longitude_bnds" ; # longitude:units = "degrees_east" ; # longitude:standard_name = "longitude" ; # double longitude_bnds(longitude, bnds) ; # double forecast_period ; # forecast_period:bounds = "forecast_period_bnds" ; # forecast_period:units = "hours" ; # forecast_period:standard_name = "forecast_period" ; # double forecast_period_bnds(bnds) ; # double forecast_reference_time(time) ; # forecast_reference_time:bounds = "forecast_reference_time_bnds" ; # forecast_reference_time:units = "hours since 1970-01-01" ; # forecast_reference_time:standard_name = "forecast_reference_time" ; # forecast_reference_time:calendar = "360_day" ; # double forecast_reference_time_bnds(time, bnds) ; # # // global attributes: # :Conventions = "CF-1.5" ; # :date = "01/01/60" ; # :emission_type = "2" ; # :history = "Tue Jun 18 14:32:42 BST 2013 - XCONV V1.92 16-February-2006" ; # :invalid_standard_name = "tendency_of_atmosphere_mass_content_of_ALICE_due_to_emission" ; # :name = "ALICE" ; # :source = "UKCA Tutorial Task 5.1 - creating netCDF emissions" ; # :time = "00:00" ; # :title = "Emissions of ALICE in kg/m^2/s" ; # :update_freq_in_hours = "120" ; # :update_type = "2" ; # :valid_max = 2.60646e-08f ; # :valid_min = 0.f ; # }
All scripts provided for these tutorials can also be found on GitHub.
Solution to Task5.1
You were given the task:
- In the
Tutorials/vn11.8/Task05.1
directory there is the file Emissions_of_ALICE.nc which is a 0.5x0.5 degree resolution surface emission field. You should regrid this file to the N48 ENDGame grid, and output it as a netCDF file that has the required netCDF metadata for a climatological surface emission without any diurnal cycle.
It should be straight-forward to point to your .pa file to use in the script. You should read through it and have an understanding of how it works.
Once the file has been regridded, you should see the output looking like this:
A file that has been produced by the above script can be found at
Tutorials/vn11.8/Task05.1/solution/ukca_emiss_ALICE.nc
Task 5.2: make the required code changes to add your emission into UKCA
TASK 5.2: You should now make the UKCA code changes to add your emission into the ALICE tracer. Use STASH code s50i304 for the diagnosic.
| Hint |
|---|
| You will need to add-in the molar mass of ALICE. You can calculate this from the mass of air and the conversion factor defined in Task 4.2. You will need to add code to allow a diagnostic of the emission to be output. |
Rose changes
There are several ways that the location of emissions files can be defined within the UKCA panel. These are:
- With a full path to the file
- Using environment variables set using an ancil_versions file
- A mixture of (1) and (2)
- By specifying a top-level directory in the
ukca_em_dirRose variable, and then paths to the required files withinukca_em_files
Within Rose you may also find that the files are in separate boxes (one for each file) or in a single long list.
The UKCA training suite uses option 1 above, specifying the full path in ukca_em_files. Note that ukca_em_dir is blank in this suite.
Make your Rose changes
It is easier to add the emissions file by editing the app/um/rose-app.conf file directly in a text editor than by using rose edit. This is because Rose can sometimes make all the files appear on a single line, whereas in the text file they will be on separate lines. To find this block save and close your suite, and then open the app/um/rose-app.conf file in a text editor (e.g. emacs, vim, or leafpad). Search for ukca_em_files and you will find the text below:
ukca_em_files='$UMDIR/ancil/atmos/n48e/ukca_emiss/gfed3.1/clim_2002_2011/v2/ukca_emiss_BC_biomass.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/gfed3.1/clim_2002_2011/v2/ukca_emiss_OC_biomass.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/andres_kasgnoc/v1/ukca_emiss_SO2_nat.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_Monoterp.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/1970_2010/v1/ukca_emiss_SO2_low.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_BC_fossil.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_BC_biofuel.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_OC_fossil.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_OC_biofuel.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/1970_2010/v1/ukca_emiss_SO2_high.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/1970_2010/v1/ukca_emiss_DMS.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/1970_2010/v1/ukca_emiss_NH3.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_NO_aircrft.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_HCHO.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_CH4.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_MeCHO.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_C3H8.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_NO.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_C2H6.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_Me2CO.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_C5H8.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_NVOC.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_CO.nc'
you will need to add a new line at the end of the block, with the = sign aligned, specifying the path to your new file. Don't forget to add a comma (,) at the end of the line above.
Code changes
ukca_config_defs_mod.F90
You will need to edit the em_chem_spec array for the scheme that you are using to include the new species that you are emitting into. By convention, these are ordered as 2D fields followed by 3D fields, with chemical emissions first and then aerosol emissions, although this order is unimportant other than the fact that the 2D emissions should be placed before the 3D emissions of 'SO2_nat', 'BC_biomass', 'OC_biomass', and 'NO_aircrft' if these are present.
If the tracer is purely chemical and is always on (i.e. is not included only when aerosol chemistry is turned on) then you will need to put this in twice. Also remember to increment this size of the em_chem_spec array, given by n_chem_emissions variable.
ukca_constants.F90
You will need to define the M_species for the emitted species. This should be consistent with the C_species value set in the previous tutorial.
get_molmass_mod.F90
You will need to add to the species_name CASE statement, to include a line such as this
CASE ('ALICE ')
get_molmass = m_ALICE
for each new species that you are emitting into.
Emissions diagnostics
By default, when emitting into a new species, the code requires you to create a diagnostic for these emissions as well. This is done by considering the following file and routines:
STASHmaster_A and STASHmaster-meta.conf
You should add a diagnostic in a similar way to the section 34 tracers, but in this case to section 50. It is best to copy an existing 2D (or 3D) emissions diagnostic specification into an empty slot at the end of the emissions list (e.g. section 50, item 315 for instance), and make only the required edits to that.
Once you have made these code changes you should commit your branch to ensure that these STASHmaster changes are picked-up by your suite correctly when running.
get_emdiag_stash_mod.F90
Once you have decided on the STASH code to use, you need to add the species and 3-digit item code to this routine in the CASE statement.
ukca_emdiags_struct_mod.F90
This routine contains the derived type that holds the emissions fluxes for all species. You will need to add a logical (used to determine whether the output has been requested in STASH) and a pointer to a 2D or 3D array that will hold the emissions flux.
ukca_update_emdiagstruct_mod.F90
In this routine you will need to add the initialisation of the logical added in ukca_emdiags_struct_mod.F90, as well as a small block of code that copies the 2D/3D em_diags data into the newly created pointer in the derived type. The logical will also need to be set to .TRUE. if STASH output has been requested (determined by the sf(item,section) look-up array).
Here is it easiest to copy an existing code-block and make the necessary changes for your new emission.
ukca_emiss_diags_mod.F90
In this routine you will add a unique code-block for this diagnostic that copies the pointer in the derived type into the stashwork array using the copydiag or copydiag_3d routines. The stashwork array contains all the fields that will be output through STASH.
Here is it easiest to copy an existing code-block and make the necessary changes for your new emission.
ukca_emiss_ctl_mod.F90
The call to ukca_emiss_diags is protected by an IF statement, and you'll need to add a check to the entry in the sf array to see if the diagnostic is on. This should be of the format sf(N,section), where N is the item number of your new emissions diagnostic.
Solution to Task 5.2
You were given the task
- You should now make the UKCA code changes to add your emission into the ALICE tracer. Use STASH code s50i304 for the diagnosic.
You were given the hints
- You will need to add-in the molar mass of ALICE. You can calculate this from the mass of air and the conversion factor defined in Task 4.2.
- You will need to add code to allow a diagnostic of the emission to be output.
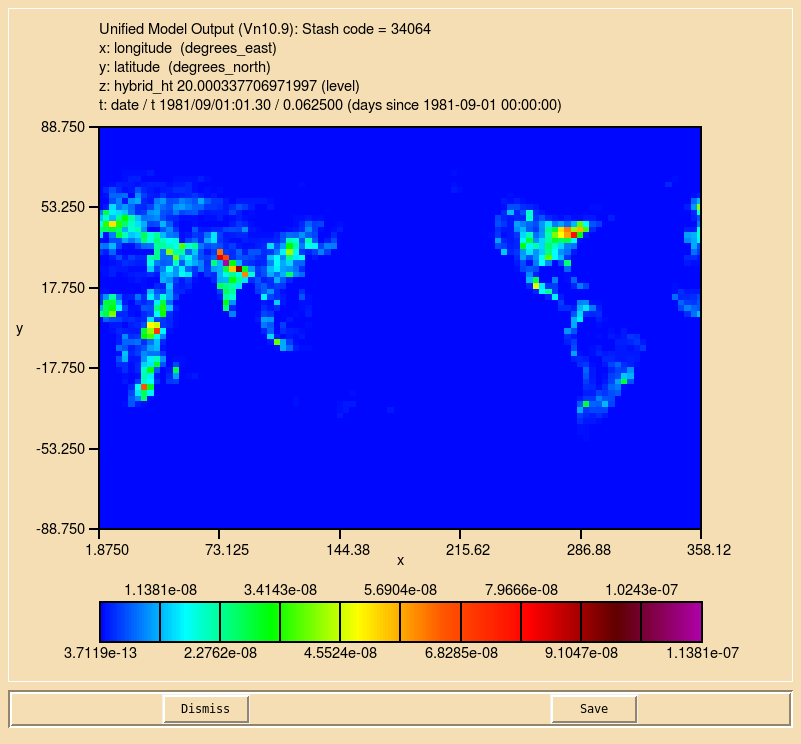
Once you have made the required code and Rose changes, you should now see that you ALICE tracer (s34i064) no longer contains noise, but now looks a bit like the emissions field you created in Task 5.1.
For a working Rose suite that has completed this task, please see
- vm:
u-ca024@181072
The specific Rose changes made are:
vm:
Index: app/um/rose-app.conf
===================================================================
--- app/um/rose-app.conf (revision 181072)
+++ app/um/rose-app.conf (revision 182181)
@@ -2924,7 +2924,8 @@
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_Me2CO.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_C5H8.nc',
='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_NVOC.nc',
- ='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_CO.nc'
+ ='$UMDIR/ancil/atmos/n48e/ukca_emiss/cmip5/2000/v1/ukca_emiss_CO.nc',
+ ='/umshared/Tutorials/vn11.8/Task05.1/solution/ukca_emiss_ALICE.nc'
ukca_h1202mmr=3.788e-13
ukca_h1211mmr=2.225e-11
ukca_h1301mmr=1.363e-11
Index: app/fcm_make/rose-app.conf
===================================================================
--- app/fcm_make/rose-app.conf (revision 181072)
+++ app/fcm_make/rose-app.conf (revision 182181)
@@ -43,4 +43,4 @@
thread_utils=false
timer_version=3A
um_rev=$BASE_UM_REV
-um_sources=branches/dev/lukeabraham/vn11.8_UKCA_Tutorial_Solns@93589
+um_sources=branches/dev/lukeabraham/vn11.8_UKCA_Tutorial_Solns@93871
These differences can be found in the file Tutorials/vn11.8/worked_solutions/Task05.2/Task05.2_rose.patch.
The specific UM changes made are:
Index: src/atmosphere/UKCA/get_molmass_mod.F90
===================================================================
--- src/atmosphere/UKCA/get_molmass_mod.F90 (revision 93589)
+++ src/atmosphere/UKCA/get_molmass_mod.F90 (revision 93871)
@@ -50,7 +50,7 @@
m_nh3, m_no, m_no2, m_oclo, m_ocs, m_oxylene, &
m_so2, m_so4, m_toluene, m_c2h2, m_tbut2ene, &
m_benzene, m_etoh, m_etcho, m_mek, m_hcooh, &
- m_meco2h, m_hoch2cho
+ m_meco2h, m_hoch2cho, m_ALICE
USE ereport_mod, ONLY: ereport
USE parkind1, ONLY: jpim, jprb ! DrHook
@@ -284,6 +284,11 @@
get_molmass = m_hoch2cho
! -----------------------------------------
+ ! UKCA Tutorial Tracer
+CASE ('ALICE ')
+ get_molmass = m_ALICE
+
+ ! -----------------------------------------
! Others (report warning)
CASE ('AGE ')
get_molmass = 1.0
Index: src/atmosphere/UKCA/ukca_constants.F90
===================================================================
--- src/atmosphere/UKCA/ukca_constants.F90 (revision 93589)
+++ src/atmosphere/UKCA/ukca_constants.F90 (revision 93871)
@@ -587,6 +587,9 @@
! - Using m_cri = 150g/mol (same as Sec_org)
REAL, PARAMETER :: m_cri = 150.0
+! UKCA Tutorial tracers - only ALICE is emitted
+REAL, PARAMETER :: m_ALICE = 28.97
+
! For solar angle calculations
REAL, PARAMETER :: fxb = 23.45/Recip_Pi_Over_180
! tropic of capricorn
Index: src/atmosphere/UKCA/ukca_emiss_diags_mod.F90
===================================================================
--- src/atmosphere/UKCA/ukca_emiss_diags_mod.F90 (revision 93589)
+++ src/atmosphere/UKCA/ukca_emiss_diags_mod.F90 (revision 93871)
@@ -498,6 +498,15 @@
emdiags%em_mek (:,:), row_length, rows)
END IF
+!---------------------------------------------------------------------
+! Sec 50, item 315: ALICE surface emissions for UKCA Tutorial
+item = get_emdiag_stash ('ALICE ')
+IF (sf(item, section)) THEN
+ CALL copydiag (stashwork (si(item,section,im_index): &
+ si_last(item,section,im_index)), &
+ emdiags%em_ALICE (:,:), row_length, rows)
+END IF
+
IF (lhook) CALL dr_hook(ModuleName//':'//RoutineName,zhook_out,zhook_handle)
RETURN
Index: src/atmosphere/UKCA/ukca_emiss_ctl_mod.F90
===================================================================
--- src/atmosphere/UKCA/ukca_emiss_ctl_mod.F90 (revision 93589)
+++ src/atmosphere/UKCA/ukca_emiss_ctl_mod.F90 (revision 93871)
@@ -1117,7 +1117,7 @@
sf(304,section) .OR. sf(305,section) .OR. sf(306,section) .OR. &
sf(307,section) .OR. sf(308,section) .OR. sf(309,section) .OR. &
sf(310,section) .OR. sf(311,section) .OR. sf(312,section) .OR. &
- sf(313,section) .OR. sf(314,section) ) THEN
+ sf(313,section) .OR. sf(314,section) .OR. sf(315,section) ) THEN
CALL ukca_emiss_diags (row_length, rows, model_levels, &
len_stashwork50, stashwork50)
Index: src/atmosphere/UKCA/ukca_config_defs_mod.F90
===================================================================
--- src/atmosphere/UKCA/ukca_config_defs_mod.F90 (revision 93589)
+++ src/atmosphere/UKCA/ukca_config_defs_mod.F90 (revision 93871)
@@ -307,7 +307,7 @@
! =======================
ELSE IF (ukca_config%l_ukca_strattrop) THEN
IF (.NOT. ukca_config%l_ukca_achem) THEN ! If NOT using aerosol chemistry
- n_chem_emissions = 10
+ n_chem_emissions = 11
n_3d_emissions = 1 ! aircraft NOX
n_aero_tracers = 0
ALLOCATE(em_chem_spec(n_chem_emissions+n_3d_emissions))
@@ -314,13 +314,13 @@
em_chem_spec = &
(/'NO ','CH4 ','CO ','HCHO ', &
'C2H6 ','C3H8 ','Me2CO ','MeCHO ', &
- 'C5H8 ', nm_meoh_em ,'NO_aircrft'/)
+ 'C5H8 ', nm_meoh_em ,'ALICE ','NO_aircrft'/)
n_chem_tracers = 73 ! No chem tracers
nr_therm = 220 ! thermal reactions
nr_phot = 55 ! photolytic (ATA)
ELSE ! If using aerosol chemistry
- n_chem_emissions = 19 ! em_chem_spec below
+ n_chem_emissions = 20 ! em_chem_spec below
n_3d_emissions = 4 ! BC, OC, volc SO2 & aircraft NOX
ALLOCATE(em_chem_spec(n_chem_emissions+n_3d_emissions))
em_chem_spec = &
@@ -327,9 +327,9 @@
(/'NO ','CH4 ','CO ','HCHO ', &
'C2H6 ','C3H8 ','Me2CO ','MeCHO ', &
'C5H8 ','BC_fossil ','BC_biofuel','OC_fossil ', &
- 'OC_biofuel','Monoterp ', nm_meoh_em ,'SO2_low ', &
- 'SO2_high ','NH3 ','DMS ','SO2_nat ', &
- 'BC_biomass','OC_biomass','NO_aircrft'/)
+ 'OC_biofuel','Monoterp ', nm_meoh_em ,'ALICE ', &
+ 'SO2_low ','SO2_high ','NH3 ','DMS ', &
+ 'SO2_nat ','BC_biomass','OC_biomass','NO_aircrft'/)
n_aero_tracers = 12
n_chem_tracers = 73 ! No chem tracers
IF (ukca_config%l_ukca_trophet) THEN
Index: src/atmosphere/UKCA/get_emdiag_stash_mod.F90
===================================================================
--- src/atmosphere/UKCA/get_emdiag_stash_mod.F90 (revision 93589)
+++ src/atmosphere/UKCA/get_emdiag_stash_mod.F90 (revision 93871)
@@ -175,6 +175,9 @@
CASE ('MEK ')
get_emdiag_stash = 314
+CASE ('ALICE ')
+ get_emdiag_stash = 315
+
CASE DEFAULT
! Report error unless this is an aerosol emission (BC_fossil:, etc)
! No diagnostics for these source emissions because the resulting
Index: src/atmosphere/UKCA/ukca_emdiags_struct_mod.F90
===================================================================
--- src/atmosphere/UKCA/ukca_emdiags_struct_mod.F90 (revision 93589)
+++ src/atmosphere/UKCA/ukca_emdiags_struct_mod.F90 (revision 93871)
@@ -65,6 +65,8 @@
LOGICAL :: l_em_etcho
LOGICAL :: l_em_hoch2cho
LOGICAL :: l_em_mek
+ ! UKCA Tutorial
+ LOGICAL :: l_em_ALICE
! Pointers to hold emission diagnostics
@@ -104,6 +106,8 @@
REAL, POINTER :: em_etcho (:,:)
REAL, POINTER :: em_hoch2cho(:,:)
REAL, POINTER :: em_mek (:,:)
+ ! UKCA Tutorial
+ REAL, POINTER :: em_ALICE (:,:)
END TYPE emdiags_struct
Index: src/atmosphere/UKCA/ukca_update_emdiagstruct_mod.F90
===================================================================
--- src/atmosphere/UKCA/ukca_update_emdiagstruct_mod.F90 (revision 93589)
+++ src/atmosphere/UKCA/ukca_update_emdiagstruct_mod.F90 (revision 93871)
@@ -123,6 +123,8 @@
emdiags%l_em_etcho = .FALSE.
emdiags%l_em_hoch2cho= .FALSE.
emdiags%l_em_mek = .FALSE.
+ ! UKCA Tutorial
+ emdiags%l_em_ALICE = .FALSE.
l_first = .FALSE.
END IF
@@ -533,6 +535,17 @@
END IF
END IF
+! UKCA Tutorial
+CASE ('ALICE ')
+ IF (emdiags%l_em_ALICE) THEN
+ emdiags%em_ALICE (:,:) = em_diags (:,:,1)
+ ELSE
+ IF (sf(item,section)) THEN
+ ALLOCATE (emdiags%em_ALICE (row_length, rows))
+ emdiags%em_ALICE (:,:) = em_diags (:,:,1)
+ emdiags%l_em_ALICE = .TRUE.
+ END IF
+ END IF
CASE DEFAULT
! Report error unless this is an aerosol emission (BC_fossil:, etc)
Index: rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster-meta.conf
===================================================================
--- rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster-meta.conf (revision 93589)
+++ rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster-meta.conf (revision 93871)
@@ -26276,6 +26276,14 @@
=
=Available only if NetCDF emission system of UKCA is used
+[stashmaster:code(50315)]
+description=ALICE surf emissions (kg m-2 s-1)
+help=Surface emission flux of ALICE (2D)
+ =kg m-2 s-1
+ =Valid on all timesteps
+ =
+ =Available only if NetCDF emission system of UKCA is used
+
[stashmaster:code(51001)]
description=O3 MASS MIXING RATIO ON PRESS LEVS
help=Ozone Mass Mixing Ratio in kg/kg(Air)
Index: rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster_A
===================================================================
--- rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster_A (revision 93589)
+++ rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster_A (revision 93871)
@@ -27515,6 +27515,12 @@
4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
5| 0 | 0 | 0 | 129 | 0 | 0 | 0 | 0 | 0 |
#
+1| 1 | 50 | 315 |ALICE surf emissions (kg m-2 s-1) |
+2| 0 | 0 | 1 | 1 | 5 | -1 | -1 | 0 | 0 | 0 | 0 |
+3| 000000000000000000000000010000 | 00000000000000000001 | 3 |
+4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
+5| 0 | 0 | 0 | 129 | 0 | 0 | 0 | 0 | 0 |
+#
#===============================================================================
# Section 50 Item 999 reserved.
#===============================================================================
These differences can be found in the file Tutorials/vn11.8/worked_solutions/Task05.2/Task05.2_code.patch.
Sample output from this task can be found at Tutorials/vn11.8/sample_output/Task05.2/atmosa.pa19810901_00.
Task 5.3: Output the diagnostic of your new emission
Task 5.3: Now that you are able to run the model with your new emission file, output this as a diagnostic to the UPA stream as a 3-hour mean.
You should reference the what is STASH tutorial for information on how to do this
Solution to Task 5.3
You were given the task:
- Now that you are able to run the model with your new emission file, output this as a diagnostic to the UPA stream as a 3-hour mean.
This achieved by outputting s50i304 to the UPA stream in STASH.
You will now find the additional field in your output file:
25 : 96 72 1 1 unspecified: Stash code 50315
For a working Rose suite that has completed this task, please see
- vm:
u-ca024@182503
The specific Rose changes made are:
The specific Rose changes made are:
vm: Index: app/um/rose-app.conf
=================================================================== --- app/um/rose-app.conf (revision 182181) +++ app/um/rose-app.conf (revision 182503) @@ -3785,6 +3785,14 @@ tim_name='T3HMN' use_name='UPA' +[namelist:umstash_streq(50315_f7d499b0)] +dom_name='DIAG' +isec=50 +item=315 +package= +tim_name='T3HMN' +use_name='UPA' + [namelist:umstash_streq(51001_3e6241a4)] dom_name='DP27CCM' isec=51
These differences can be found in the file Tutorials/vn11.8/worked_solutions/Task05.3/Task05.3_code.patch.
Sample output from this task can be found at Tutorials/vn11.8/sample_output/Task05.3/atmosa.pa19810901_00.
Checklist
- ☐ Obtain emissions data for the species of interest.
- ☐ Regrid the emissions to the correct MetUM resolution that you are using.
- ☐ Save these emissions as netCDF, including the required metadata that UKCA requires.
- ☐ In your suite, include this file in the
ukca_em_filesvariable in the UKCA panel: um namelist UM Science Settings Section 34 - UKCA: UK Aerosols and Chemistry. - ☐ (Optional) Put a 1 in the E column of the
chch_defs_masterinukca_chem_master.F90. - ☐ Put the species being emitted into the
em_chem_specarray inukca_setd1defs.F90, and incrementn_chem_emissions. - ☐ Put the correct M_species value in
ukca_constants.F90. - ☐ In
get_molmass_mod.F90, append the CASE statement with your new species. - ☐ Make diagnostics output slots for your new emissions in
STASHmaster_Aand add help-text toSTASHmaster-meta.conf. - ☐ In
get_emdiag_stash_mod.F90add the species and 3-digit item code to the CASE statement. - ☐ In
ukca_emdiags_struct_mod.F90add a logical and array pointer to the derived type for your new species. - ☐ In
ukca_update_emdiagstruct_mod.F90initialise your new derived type entries, and add a block of code to copy the emissions field into the diagnostic pointer. - ☐ In
ukca_emiss_diags_mod.F90add a code block to copy the field to thestashworkarray usingcopydiag/copydiag_3d. - ☐ In
ukca_emiss_ctl_mod.F90add thesf(ITEM,section)check in the IF statement protecting the call toukca_emiss_diags. - ☐
fcm committhe changes to your branch. - ☐ Output your emissions diagnostic in: um namelist Model Input and Output STASH Requests and Profiles STASH Requests.
- ☐ Run the TidyStashTransform transform macro.
- ☐ Save your suite.
- ☐ In the
roses/[SUITE-ID]directory, runfcm committo commit your changes to the repository. - ☐ Run your suite.
Written by Luke Abraham 2020. Many thanks to Alistair Sellar for the notes on netCDF metadata.