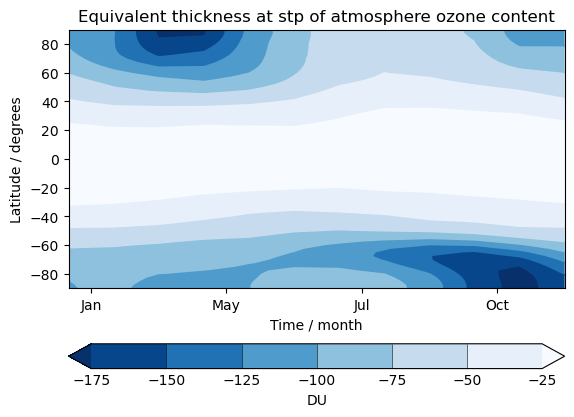
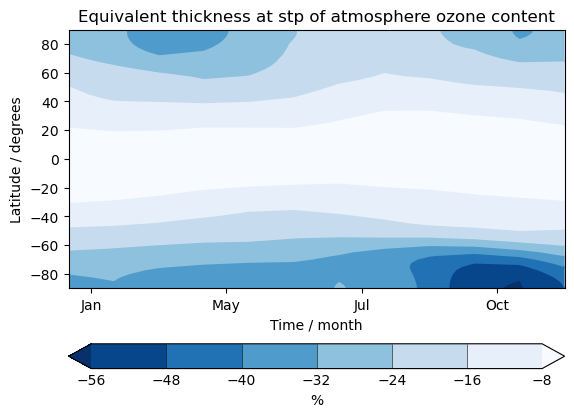
UKCA Chemistry and Aerosol UMvn13.0 Tutorial 13
UKCA Chemistry and Aerosol Tutorials at UMvn13.0
| Difficulty | HARD |
| Time to Complete | 2 or more hours |
| Video instructions |
What you will learn in this Tutorial
In this tutorial you will learn how to plot and process UKCA data using the Iris and cf python packages. Example notebooks are provided using both packages, which can be also be viewed here:
and can be found in the
Tutorials/UMvn13.0/notebooks
directory on your virtual machine.
You should work through the notebook to see how the plots are generated. Also compare the different methods between cf and Iris:
- Are some thing easier or harder to do using a particular package?
- Does one package seem to do some things better or worse than the other?
At the end of each notebook are some suggested examples which you should think about and complete.
Data
There are a number of different datasets in use in this Tutorial, all of which are Total Column Ozone (TCO). While all the data is gridded, there are a range of resolutions and file types used.
Observations
This tutorial also makes use of the Bodeker Scientific version 3.4.1 TCO dataset. For ease of use this has been processed from annual files into a single file using the following commands:
for i in `ls *.nc`; do echo $i; ncks -O --mk_rec_dmn time $i $i; done for i in `ls *.nc`; do echo $i; ncatted -O -a created,global,d,, $i; done for i in `ls *.nc`; do echo $i; ncatted -O -a units,longitude,o,c,degrees_east $i; done for i in `ls *.nc`; do echo $i; ncatted -O -a units,latitude,o,c,degrees_north $i; done ncrcat BSFilledTCO_V3.4.1_????_Monthly.nc ../BSFilledTCO_V3.4.1_Monthly.nc
We would like to thank Bodeker Scientific, funded by the New Zealand Deep South National Science Challenge, for providing the combined NIWA-BS total column ozone database.
Model Output
The NERC ACSIS Project
The ACSIS project was a long-term, multi-centre project funded by NERC and as part of this we produced a series UKCA simulations. Model output from one of these simulations is provided in monthly-mean UM "pp" file format. The model configuration is an N96eL85 UKESM1-AMIP (atmosphere-only) configuration at UM version 11.5, where UM winds and temperatures were relaxed to the ERA5 reanalysis dataset. The chemistry scheme used was the Stratosphere-Troposphere scheme that you have been working with in this tutorial. This model simulation used a Gregorian calendar.
Phase 1 of the Chemistry-Climate Model Initiative
UKCA output is submitted to a number of different international model inter-comparison projects, including the first phase of the Chemistry-Climate Model Initiative (CCMI1). Here we submitted data from an atmosphere-only simulation produced using UKCA run at UM version 7.3 at N48L60 resolution used a stratospheric chemistry scheme with explicit treatment of CFCs. Output was processed and converted to CF-standard NetCDF files. As with most UM climate simulations, this model used a 360-day calendar, where each month has 30 days.
Chemistry-Climate Model Initiative 2022
An improved version of UKESM1 was developed which reduced some of the high ozone biases, and was used to perform simulations for the [ https://blogs.reading.ac.uk/ccmi/ccmi-2022/CCMI2022 project]. Here we submitted data from three atmosphere-only UKESM1-AMIP simulations (with to different initial conditions but other settings and forcings the same) run at UM version 11.1 at N96eL85 resolution and uses the improved Stratosphere-Troposphere chemistry and associated settings. Output was processed and converted to CF-standard NetCDF files. As with most UM climate simulations, this model used a 360-day calendar, where each month has 30 days.
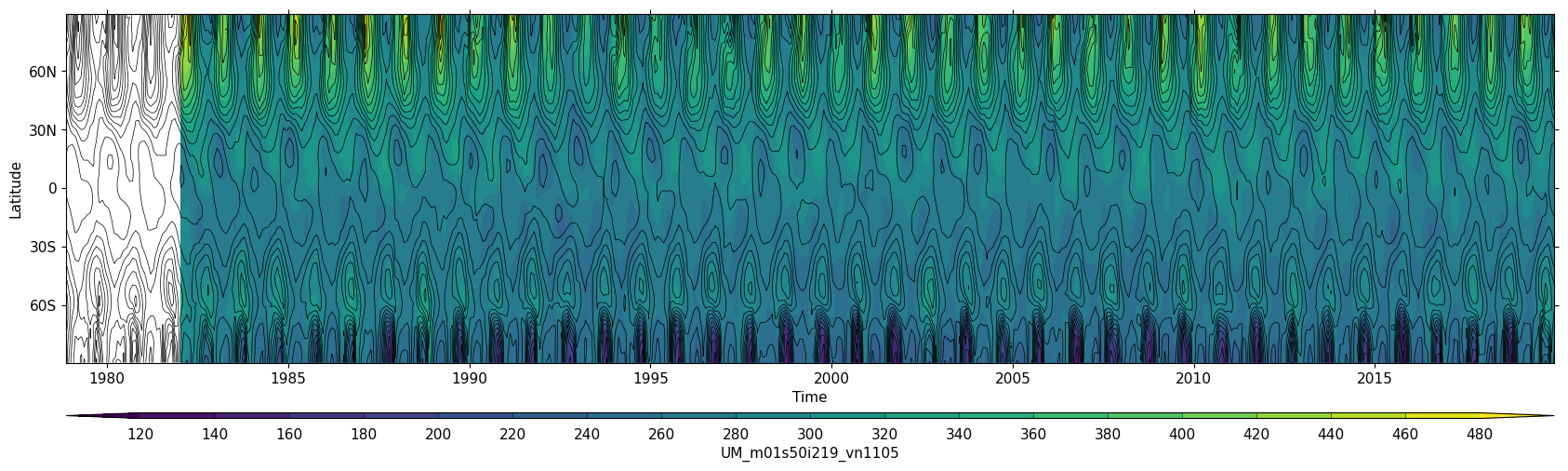
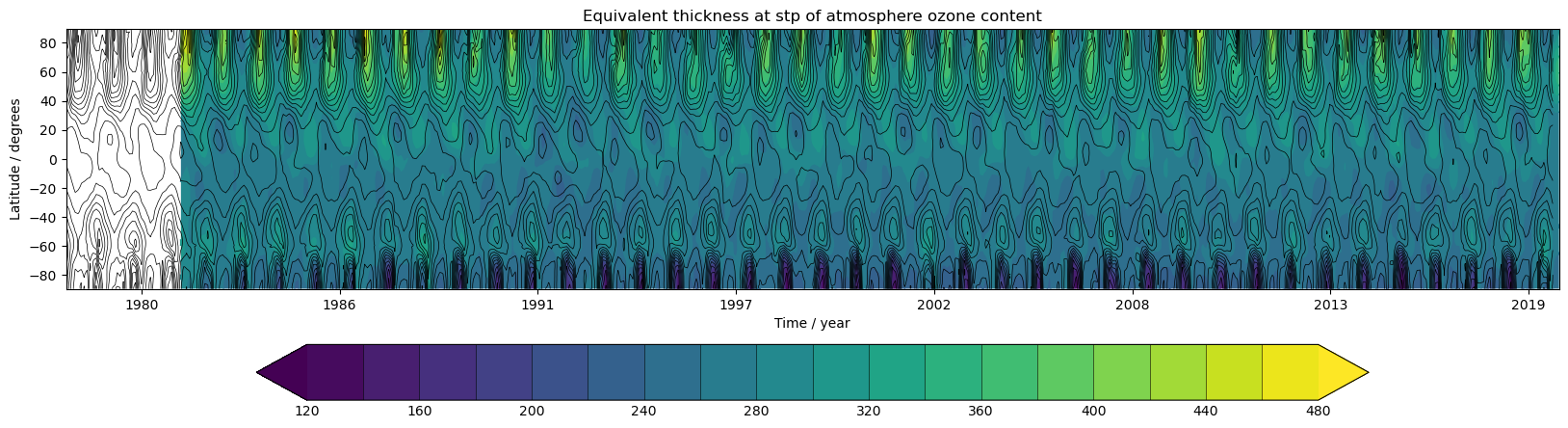
Exercise 1: Plot zonal-mean total column ozone
You should use the *_toz01.ipynb files for cf and for Iris:
Further exercises
When you have completed this notebook, consider the following exercises to try:
- Plot members from the CCMI2022 ensemble
- Produce a plot of the ensemble mean of the 3 ensemble members. How would you go about calculating the ensemble mean?
| Hint |
|---|
| You may need to delete the auxillary coordinates 'forecast_period' and 'forecast_reference_time' in the CCMI2022 ensemble using the cf-python 'f.del_construct()' operation. |
Solutions to these exercises can be found here:
Exercise 2: Plot the regional time evolution of total column ozone
You should use the *_toz02.ipynb files for cf and for Iris:
Further exercises
When you have completed this notebook, consider the following exercises to try:
- Try plotting different months and regions, e.g. March for 60N to 90N
- Try plotting annual mean TCO from 60S to 60N. How would you go about producing an annual mean of each year?
| Hint |
|---|
| Take a look at how to perform statistical collapses in cf. |
| Take a look at the aggregated_by() method in Iris. |
Solutions to these exercises can be found here:
- March 60N to 90N:
- Annual mean 60S to 60N
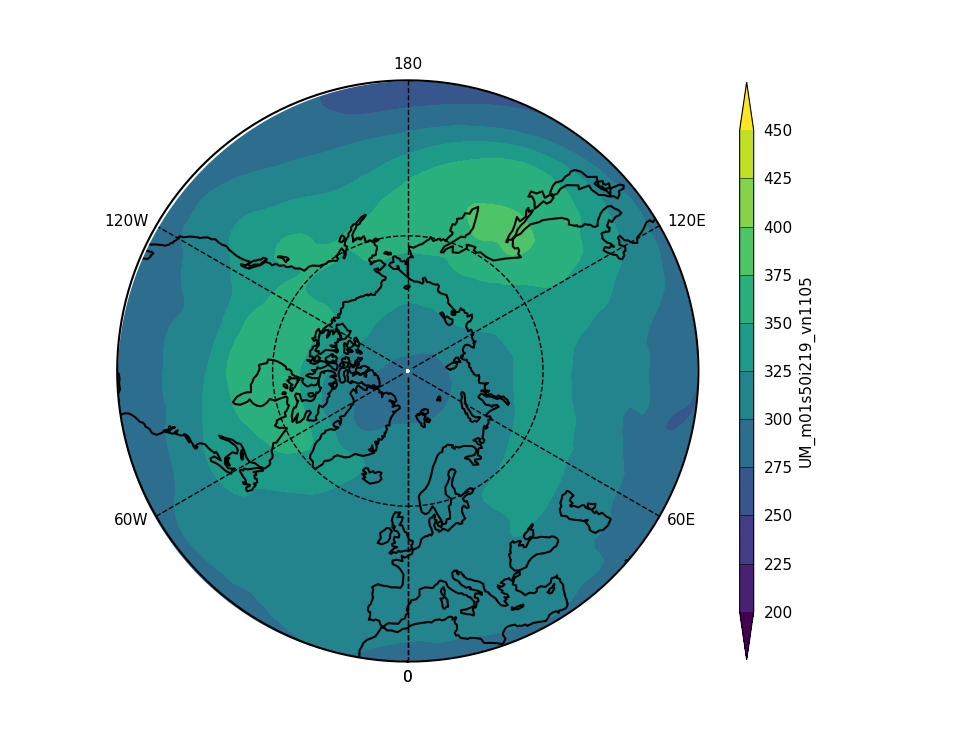
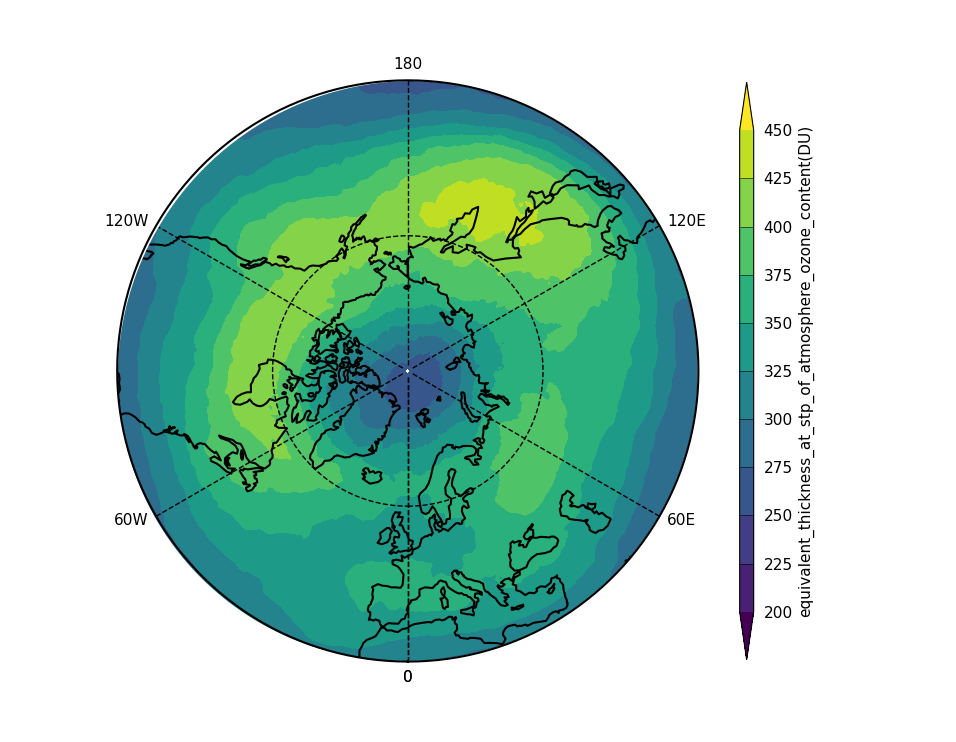
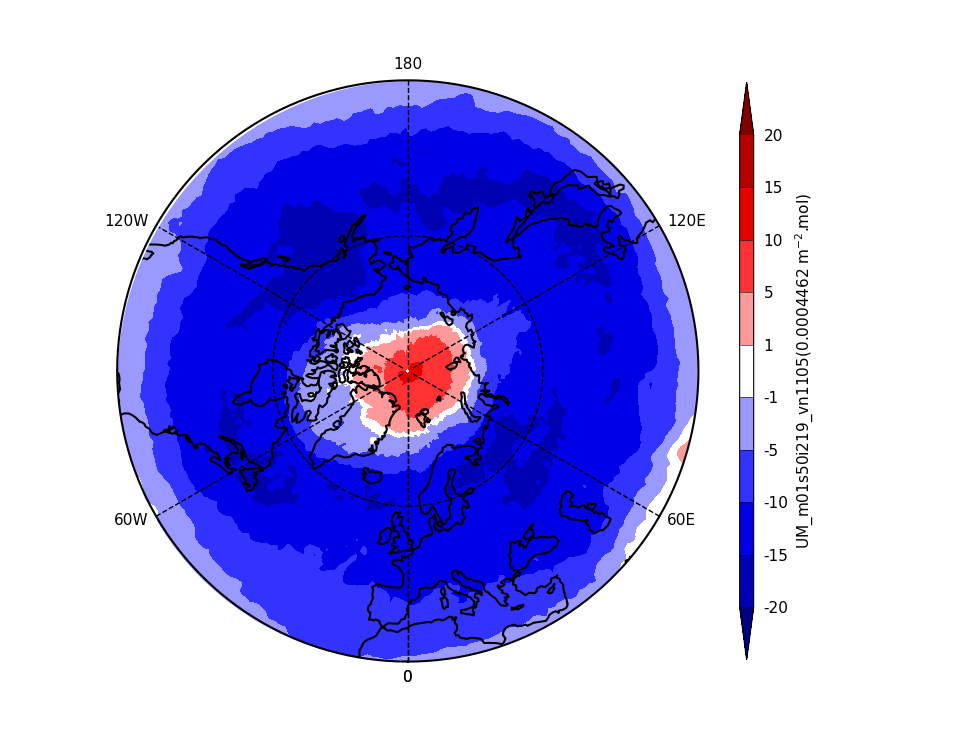
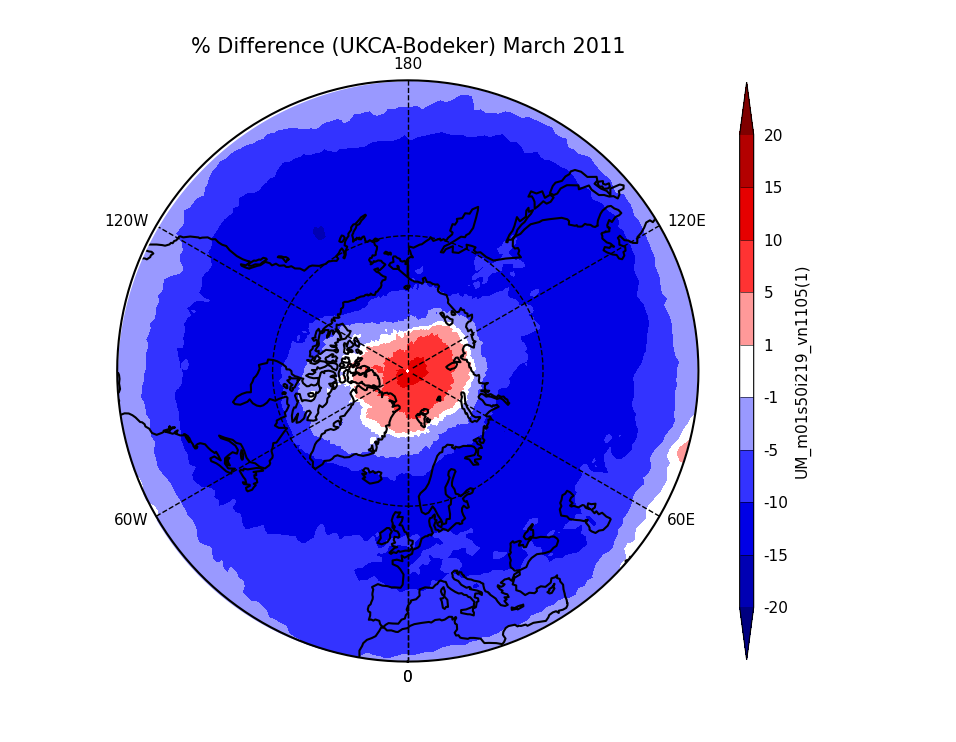
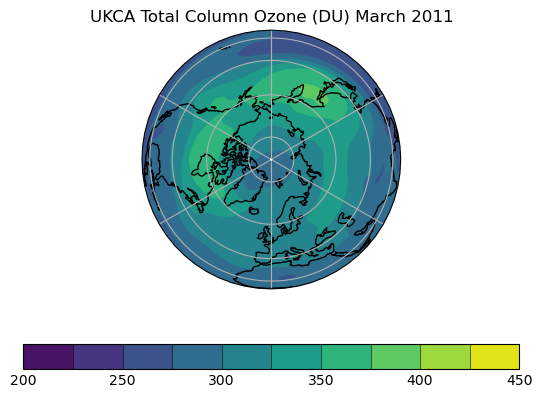
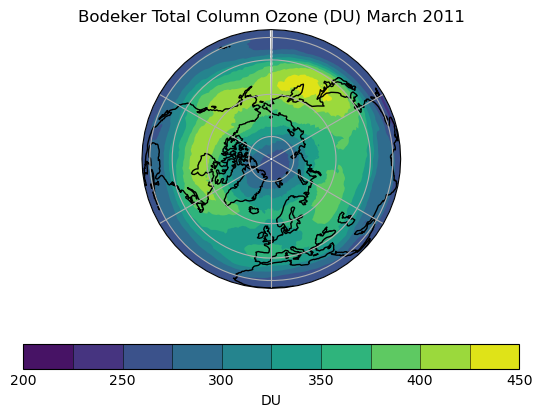
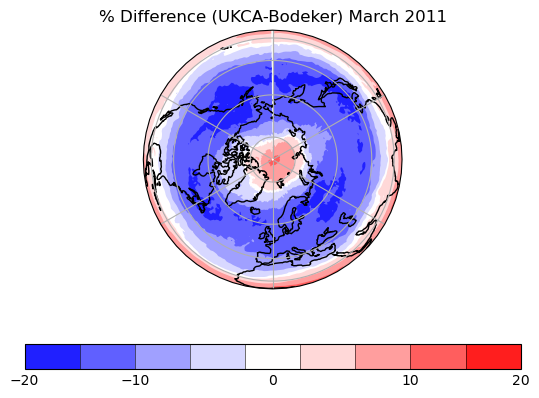
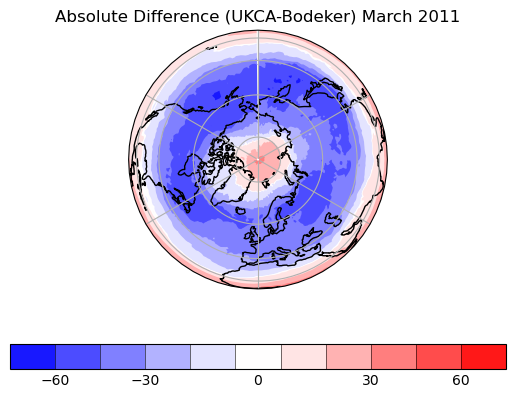
Exercise 3: Plot comparisons of total column ozone for specific months
You should use the *_toz03.ipynb files for cf and for Iris:
Further exercises
When you have completed this notebook, consider the following exercises to try:
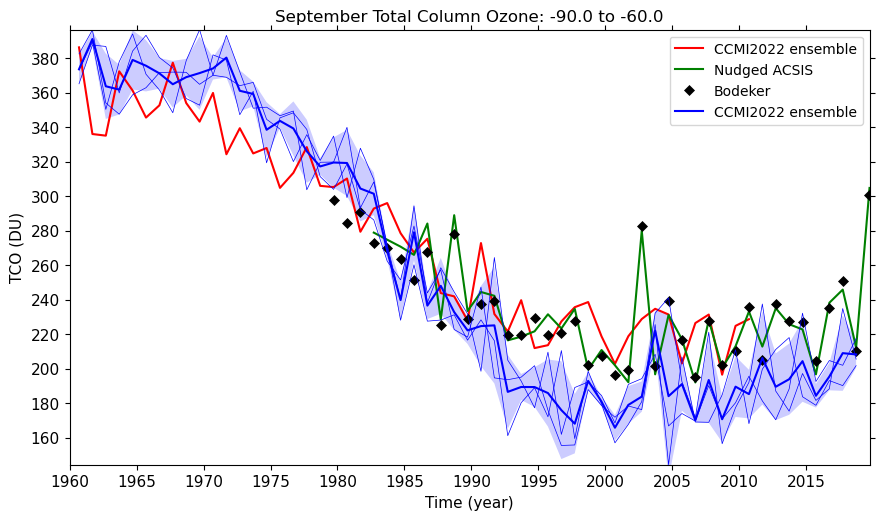
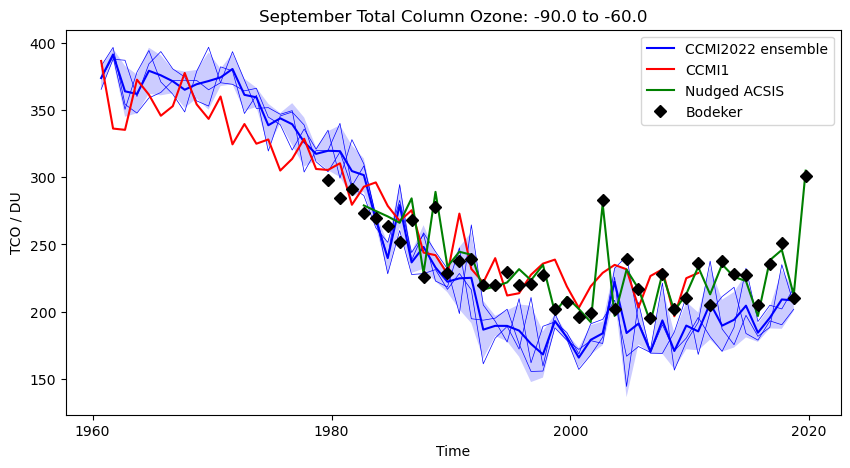
- Try plotting different dates, e.g. September 2002 over the south pole
- How do the CCMI1 and CCMI2022 similations compare. Rather than individual months, try considering decadal climatologies and compare ensemble members.
Solutions to these exercises can be found here:
- September 2002 over the south pole
- CCMI plots
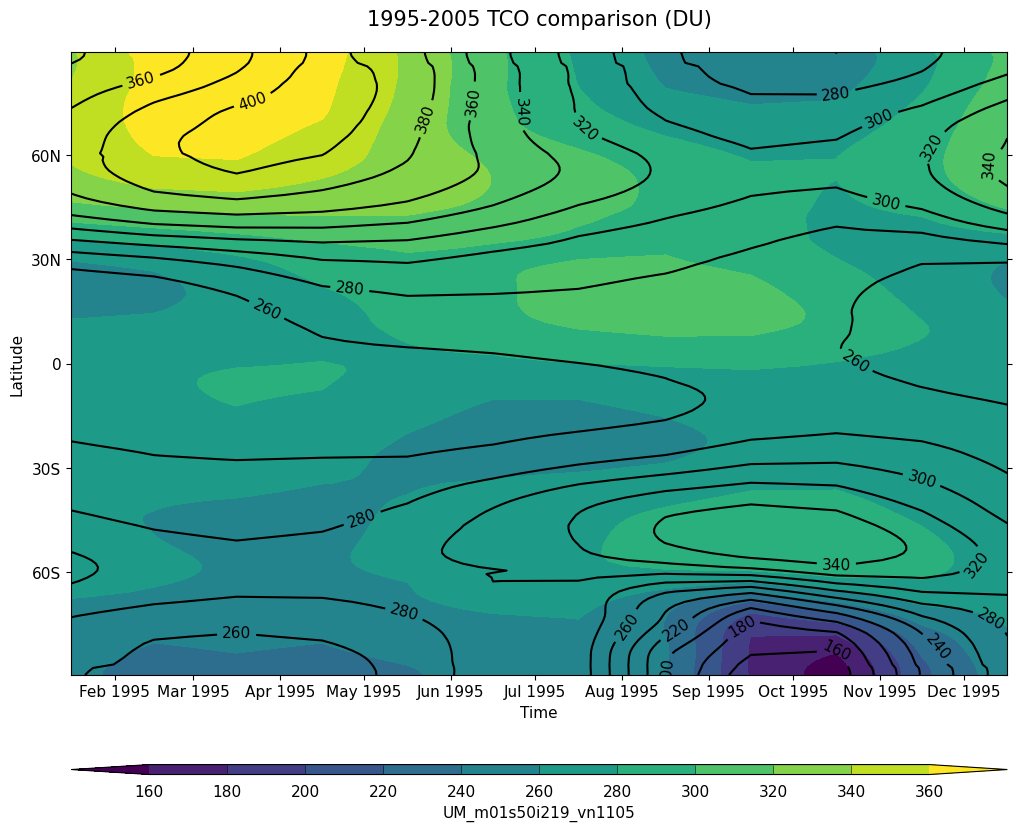
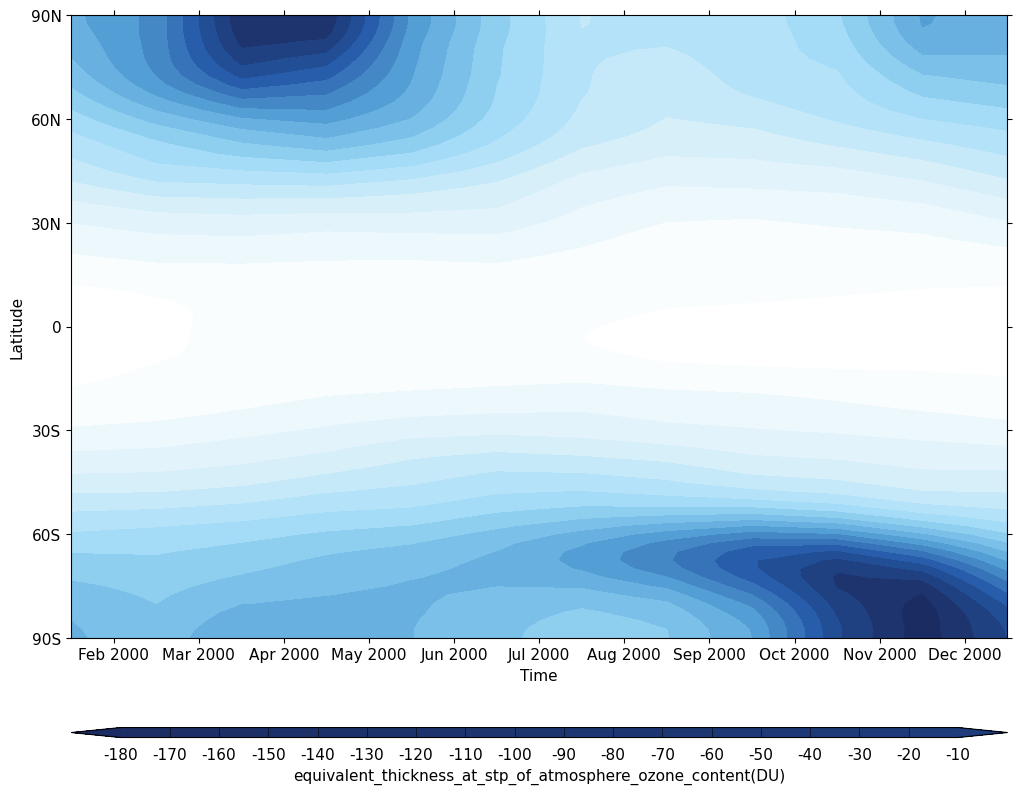
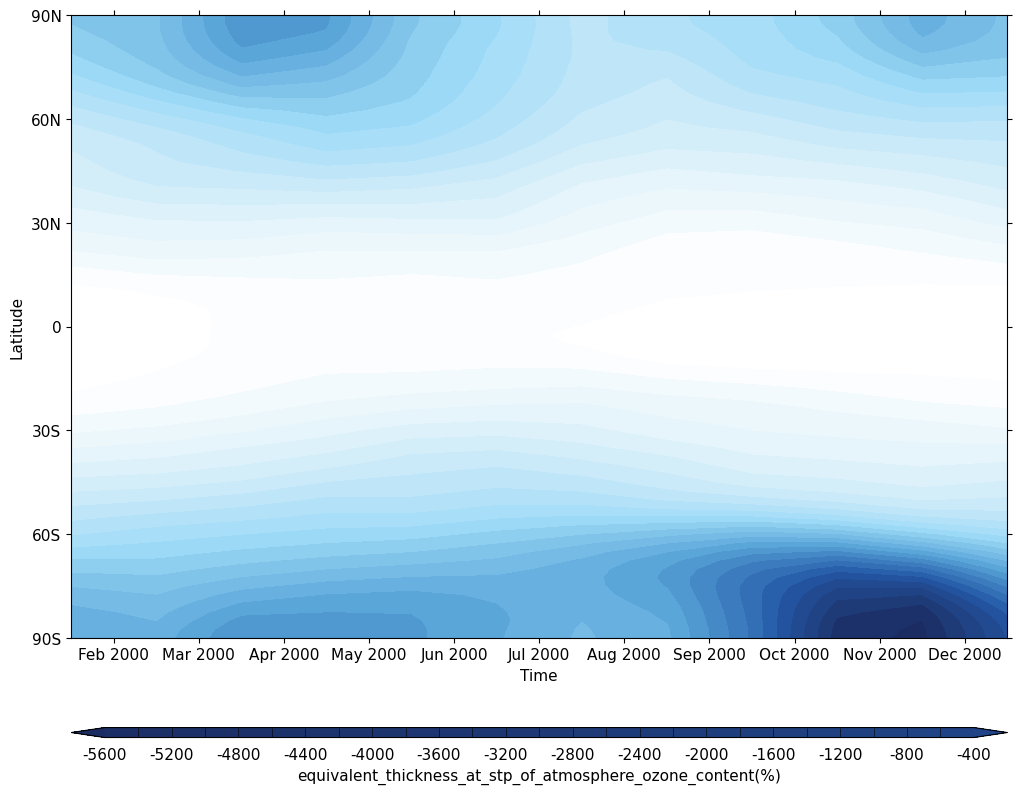
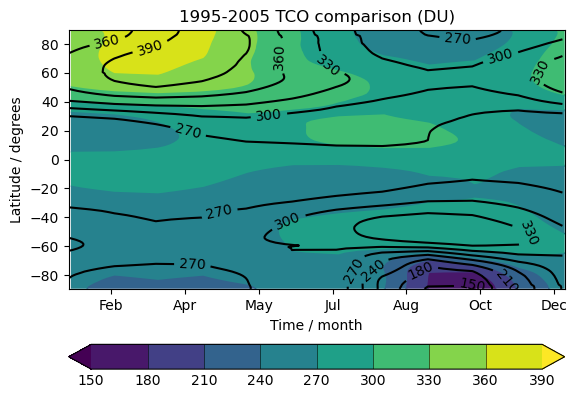
Exercise 4: Plot climatological zonal-mean total column ozone
You should use the *_toz04.ipynb files for cf and for Iris:
Further exercises
When you have completed this notebook, consider the following exercises to try:
- Try comparing the CCMI2022 ensemble members against the ensemble mean.
- Try comparing the nudged UM-UKCA ACSIS data against the CCMI2022 ensemble mean. Here the principal difference between the simulations is the use of nudging to constrain the winds and temperatures.
Checklist
- ☐ Use cf-python or Iris to process and plot model and observational data
UKCA Chemistry and Aerosol Tutorials at UMvn13.0
Written by Luke Abraham 2022.