UKCA Chemistry and Aerosol UMvn13.0 Tutorial 11: Difference between revisions
| Line 104: | Line 104: | ||
<ol start="1"> |
<ol start="1"> |
||
<li> Define your diagnostic specification blocks for the new fluxes of interest. You will need to define different blocks for each condition where the reaction changes, e.g. whether you are using aerosol chemistry or not, or following an update at a different chemistry version. This is done prior to the <tt>asad_load_default_fluxes</tt> routine. You should give these blocks consistent names, e.g. <tt>ukca_tutorial_fluxes</tt>, <tt>ukca_tutorial_fluxes_achem</tt> etc.</li> |
<li> Define your diagnostic specification blocks for the new fluxes of interest. You will need to define different blocks for each condition where the reaction changes, e.g. whether you are using aerosol chemistry or not, or following an update at a different chemistry version. This is done prior to the <tt>asad_load_default_fluxes</tt> routine. You should give these blocks consistent names, e.g. <tt>ukca_tutorial_fluxes</tt> (when not forming <tt>Sec_Org</tt>), <tt>ukca_tutorial_fluxes_achem</tt> (when forming <tt>Sec_Org</tt>) etc.</li> |
||
</ol> |
</ol> |
||
TYPE(asad_flux_defn), PARAMETER, PUBLIC :: ukca_tutorial_fluxes_achem(3) = [ & |
TYPE(asad_flux_defn), PARAMETER, PUBLIC :: ukca_tutorial_fluxes_achem(3) = [ & |
||
Revision as of 16:20, 7 December 2022
UKCA Chemistry and Aerosol Tutorials at UMvn13.0
| Difficulty | HARD |
| Time to Complete | 2 or more hours |
| Video instructions |
Remember to run mosrs-cache-password.
What you will learn in this Tutorial
In this tutorial you will learn about the UKCA diagnostics package and the different diagnostics that you can output using it. You will also learn how to add new diagnostics from the new reactions and deposition that you have added.
Task 11.1: Output new diagnostics
TASK 11.1: Output diagnostics of the reaction to STASH code 50996, the dry deposition of ALICE to STASH code 50997, and the wet deposition of BOB to 50998. They should be outputted as a 3-hour mean to the pa/UPA stream.
Adding New UKCA Diagnostics
If you are using one of the chemistry schemes that uses ASAD (e.g. TropIsop, Strat, StratTrop, or CRI-Strat) then you can make use of the ASAD Reaction Flux Diagnostics module (held in src/control/core/diagnostics/asad_flux_dat.F90). These allow you to straight-forwardly output new reaction and deposition fluxes.
To output new diagnostics you will first need to define them in the asad_flux_dat.F90 module, and then create new STASHmaster file specifications for them, as you have already done in tutorial 7.
During this tutorial you will be tasked with outputting the reaction and deposition fluxes that you have added. You will need to make changes to both your UKCA and your UM branches.
src/control/core/diagnostics/asad_flux_dat.F90
All ASAD-based chemistry schemes output their diagnostics using the asad_flux_dat.F90 module. Here the diagnostics are defined in blocks with the format
asad_flux_defn('Diagnostic type',STASH code,'Diagnostic specification',Mask,Reaction number,Number of species, & (/'Species/Reactant 1','Reactant 2'/), & (/'Product 1','Product 2','Product 3','Product 4'/)), &
Which have the following meaning:
Diagnostic Type
This is a three character string which defines what type of diagnostic is being requested. This can take the values
- RXN to output the flux through a reaction (in moles/gridcell/s)
- DEP to output the deposition flux of a species (in moles/gridcell/s)
- NET to output the net chemical tendency of a species (in moles/gridcell/s)
- STE to output the net dynamical tendency of a species (in moles/gridcell/s)
- MAS to output the mass of the atmosphere (in kg/gridcell)
- PSC to output polar stratospheric cloud diagnostics (1 when the gridcell contains a PSC, 0 otherwise - monthly mean field will be a fraction in range 0 → 1)
- TPM to output the tropospheric mask (1 for troposphere, 0 otherwise - monthly mean field will be a fraction in range 0 → 1)
- OUT to output a tracer in mmr. Only really useful if the field is masked to give the tropospheric concentration only (see the discussion of the Mask option)
STASH Code
This is a 5 digit integer defining the STASH code that the diagnostic will be outputted to (e.g. 50001). Currently this must be in section 50.
Diagnostic Specification
This is a one character string which is needed to further define what diagnostic is required, and can be different depending on Diagnostic Type (3-character string). If it isn't needed then it should just be set to X or left blank.
- RXN
- B to output the flux through a bimolecular reaction
- T to output the flux through a termolecular reaction
- H to output the flux through a heterogeneous reaction
- J to output the flux through a photolysis reaction
- DEP
- D to output the dry deposition flux
- W to output the wet deposition flux
- PSC
- 1 to output the fraction of Type 1 PSCs
- 2 to output the fraction of Type 2 PSCs
Mask
This is a logical which defines whether only the tropospheric values of the diagnostic are outputted (.TRUE.) or not (.FALSE.). It is calculated every timestep.
For the STE diagnostic this is required if you wish to output the diagnosed stratosphere-troposphere exchange of a species. For the OUT diagnostic this can be used to output only the tropospheric concentration of a tracer. This is also used in the calculation of the of the TPM diagnostic.
Reaction number
This is an integer, and should only be used in the special case of there being two (or more) reactions with the exactly the same reactants and products, but with different rate coefficients. In this case the first reaction in the list would be given number 1 and the second 2 etc. If this is not needed then it should be set to 0 (which will be usual for most reactions).
Number of Species
This is an integer, and should give the total number of species, so this will be 1 for diagnostics such as DEP, STE, NET etc., which only consider a single species, and the total number of reactants and products for the RXN diagnostic.
Species
This is a 10-character string giving the exact name of the species that the diagnostic should be considered for (including capitalisation). This is only used for the DEP, NET, STE, and OUT. For the RXN diagnostics the full list of reactants and products should be given (see below). For the MAS, PSC, and TPM diagnostics this isn't needed and could either be set to XXX or left blank. If it is needed the other reactant/product slots should be left blank.
Reactants and Products
These are 10-character strings, and should be as the reaction is defined in the src/science/core/chemistry/ukca_chem_master.F90 module.
Summing Diagnostics
If you define more than one diagnostic to be output to the same STASH code, then the diagnostic routines will sum these diagnostics together. This can be useful (e.g., if you wanted to output the sum of all NO+RO2 reactions to one STASH item), but can be problematic if you accidentally output two fields to the same STASH code, as this will give strange results!
Changes to asad_flux_dat.F90
After you have defined your new diagnostics in this module, you will need to make sure that they have been added correctly to the asad_chemical_fluxes array, which is defined in the asad_load_default_fluxes subroutine held in the asad_flux_dat.F90 module, with a size given by the n_chemical_fluxes parameter.
As there are several different chemical mechanism with further-sub options, how asad_chemical_fluxes is defined is rather complex. This is done in 6 steps:
- Define your diagnostic specification blocks for the new fluxes of interest. You will need to define different blocks for each condition where the reaction changes, e.g. whether you are using aerosol chemistry or not, or following an update at a different chemistry version. This is done prior to the asad_load_default_fluxes routine. You should give these blocks consistent names, e.g. ukca_tutorial_fluxes (when not forming Sec_Org), ukca_tutorial_fluxes_achem (when forming Sec_Org) etc.
TYPE(asad_flux_defn), PARAMETER, PUBLIC :: ukca_tutorial_fluxes_achem(3) = [ &
asad_flux_defn('RXN',50996,'B',.FALSE.,0,4, &
['ALICE ','OH '], &
['BOB ','Sec_Org ',' ',' ']), &
asad_flux_defn('DEP',50997,'D',.FALSE.,0,1, &
['ALICE ',' '], &
[' ',' ',' ',' ']), &
asad_flux_defn('DEP',50998,'W',.FALSE.,0,1, &
['BOB ',' '], &
[' ',' ',' ',' ']) &
]
TYPE(asad_flux_defn), PARAMETER, PUBLIC :: ukca_tutorial_fluxes(3) = [ &
asad_flux_defn('RXN',50996,'B',.FALSE.,0,3, &
['ALICE ','OH '], &
['BOB ',' ',' ',' ']), &
asad_flux_defn('DEP',50997,'D',.FALSE.,0,1, &
['ALICE ',' '], &
[' ',' ',' ',' ']), &
asad_flux_defn('DEP',50998,'W',.FALSE.,0,1, &
['BOB ',' '], &
[' ',' ',' ',' ']) &
]
- In asad_load_default_fluxes you will need to define an allocated array to hold the correct diagnostic specification given the various options (e.g. aerosol chemistry or not, etc.). By convention, this is prepended with aa_, e.g. aa_ukca_tutorial_fluxes.
TYPE(asad_flux_defn), ALLOCATABLE, SAVE :: aa_ukca_tutorial_fluxes(:)
- You then need to fill your aa_ array given the various possible options and for each chemistry scheme the diagnostics are valid for. You may need to define additional constants, e.g. setting a parameter ichem_version_131=131 in the example here.
! aa_ukca_tutorial_fluxes
IF (ukca_config%i_ukca_chem_version >= ichem_version_131) THEN
IF (ukca_config%l_ukca_chem_aero) THEN
ALLOCATE(aa_ukca_tutorial_fluxes(SIZE(ukca_tutorial_fluxes_achem)))
aa_ukca_tutorial_fluxes = ukca_tutorial_fluxes_achem
ELSE
ALLOCATE(aa_ukca_tutorial_fluxes(SIZE(ukca_tutorial_fluxes)))
aa_ukca_tutorial_fluxes = ukca_tutorial_fluxes
END IF
END IF
- You then need to increment n_chemical_fluxes by the size of this aa_ array (which is then used to allocate the asad_chemical_fluxes array).
IF (ALLOCATED(aa_ukca_tutorial_fluxes)) & n_chemical_fluxes = n_chemical_fluxes + SIZE(aa_ukca_tutorial_fluxes)
- You then need to copy your aa_ array into the master asad_chemical_fluxes. This is done in the way below to ensure that the entries slot into the correct place in the array, and is only done if the corresponding aa_ has been allocated (and therefore has entries in it).
IF (ALLOCATED(aa_ukca_tutorial_fluxes)) THEN p2 = p1 + SIZE(aa_ukca_tutorial_fluxes) - 1 asad_chemical_fluxes(p1:p2) = aa_ukca_tutorial_fluxes(:) p1 = p2 + 1 END IF
- You should then deallocate your aa_ array in the reverse order that they were allocated in.
IF (ALLOCATED(aa_ukca_tutorial_fluxes)) DEALLOCATE(aa_ukca_tutorial_fluxes)
This is rather complex, but this creates an extensible method of adding new diagnostics to the many different and complex options available.
Other types of diagnostics
You will need to edit the src/control/core/diagnostics/asad_chem_flux_diags.F90 module if you want to output a new type of diagnostic. This can be quite involved, but you can look at existing routines to see how things are done. You will also need to add code into the main UKCA routines the pass the data through, e.g.
! 3D flux diagnostics
IF (L_asad_use_chem_diags .AND. &
((L_asad_use_flux_rxns .OR. L_asad_use_rxn_rates) .OR. &
(L_asad_use_wetdep .OR. L_asad_use_drydep))) &
CALL asad_chemical_diagnostics(row_length,rows, &
model_levels,dpd_full,dpw_full,prk_full,y_full, &
j,i,klevel,volume,ierr)
STASHmaster file
While the diagnostics are defined in asad_flux_dat.F90 they are turned on by requesting the item through STASH. To do this you will need to edit the STASHmaster_A file in your branch, as covered in the Adding new chemical tracers to the UM tutorial. Here, as you are adding new diagnostics to section 50, the equivalent pressure-level diagnostics need to be made in section 52.
Most UKCA diagnostics are 3D, although some, such as emissions (which are outputted in a different way), are 2D. You should take care with the STASH settings in STASHmaster_A between these types, as there some differences that will need to be considered. Note that any 2D (i.e. surface) diagnostics that you create will not have a pressure-level equivalent. You will also need to add the help entries to the STASHmaster-meta.conf file.
As previously, you may find it easier to copy an existing diagnostic and edit that, but take care to ensure that the settings used are equivalent, e.g. you should copy a 3D section 50 field and not a 2D one, or one from section 34 etc. The various numbers and settings in the STASHmaster_A file define lots of different things, such as the levels used, the time validity etc. Be sure to copy an existing STASHmaster entry of the same type of field to ensure compatibility.
Remember to set the option codes on the third line as required for the diagnostics that you are adding.
Rose Changes
If you have not done so, you will also need to make sure that you use your new STASHmaster_A file in Rose, as is explained in detail in Tutorial 7.
After you have made your STASmaster_A file changes, you will need to add these diagnostics into STASH, as per Tutorial 6. Remember to run the TidyStashTransform macro. As you will have included your branch's STASHmaster_A file without a revision or using @HEAD, you won't need to make any further changes to Rose, but you will need to make sure that you have committed your UM branch prior to running. Because the file is taken from fcm:um.xm_br you will need to make sure that the revision has synced to the PUMA mirror (i.e. by waiting a few minutes).
Worked Solution to Task 11.1: Output new diagnostics
You were given the task
- Output diagnostics of the reaction to STASH code 50996, the dry deposition of ALICE to STASH code 50997, and the wet deposition of BOB to 50998. They should be outputted as a 3-hour mean to the pa/UPA stream.
The specific Rose changes made are:
Index: app/um/rose-app.conf =================================================================== --- app/um/rose-app.conf (revision 237703) +++ app/um/rose-app.conf (revision 237899) @@ -3965,6 +3965,30 @@ tim_name='T3HMN' use_name='UPA' +[namelist:umstash_streq(50996_26eab91c)] +dom_name='DALLTH' +isec=50 +item=996 +package= +tim_name='T3HMN' +use_name='UPA' + +[namelist:umstash_streq(50997_e01cc6bc)] +dom_name='DALLTH' +isec=50 +item=997 +package= +tim_name='T3HMN' +use_name='UPA' + +[namelist:umstash_streq(50998_01fd6a94)] +dom_name='DALLTH' +isec=50 +item=998 +package= +tim_name='T3HMN' +use_name='UPA' + [namelist:umstash_streq(51001_3e6241a4)] dom_name='DP27CCM' isec=51 @@ -4013,6 +4037,30 @@ tim_name='T3HMN' use_name='UPA' +[namelist:umstash_streq(52996_56c5ed16)] +dom_name='DP27CCM' +isec=52 +item=996 +package= +tim_name='T3HMN' +use_name='UPA' + +[namelist:umstash_streq(52997_eb3669f6)] +dom_name='DP27CCM' +isec=52 +item=997 +package= +tim_name='T3HMN' +use_name='UPA' + +[namelist:umstash_streq(52998_c5c157df)] +dom_name='DP27CCM' +isec=52 +item=998 +package= +tim_name='T3HMN' +use_name='UPA' + [namelist:umstash_time(t3hmn_039ecafe)] !!iedt=0 iend=-1
These differences can be found here:
Tutorials/UMvn13.0/worked_solutions/Task11.1/Task11.1_rose.patch
The specific UM changes made are:
Index: rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster-meta.conf
===================================================================
--- rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster-meta.conf (revision 112818)
+++ rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster-meta.conf (revision 112862)
@@ -27356,6 +27356,18 @@
=if interactive cloud pH scheme not used then default pH of 5 is used at all
=locations ([H+] = 1.0e-5)
+[stashmaster:code(50996)]
+description=RXN FLX: ALICE+OH->BOB(+Sec_Org)
+help=Flux through the ALICE+OH->BOB(+Sec_Org) reaction
+
+[stashmaster:code(50997)]
+description=DRY DEP FLUX: ALICE (3D)
+help=Dry deposition flux of ALICE
+
+[stashmaster:code(50998)]
+description=WET DEP FLUX: BOB (3D)
+help=Wet deposition flux of BOB
+
[stashmaster:code(51001)]
description=O3 MASS MIXING RATIO ON PRESS LEVS
help=Ozone Mass Mixing Ratio in kg/kg(Air)
@@ -31143,6 +31155,21 @@
=if interactive cloud pH scheme not used then default pH of 5 is used at all
=locations ([H+] = 1.0e-5)
+[stashmaster:code(52996)]
+description=RXN FLX: ALICE+OH->BOB PLEVS
+help=Flux through the ALICE+OH->BOB(+Sec_Org) reaction
+ = on Pressure Levels
+
+[stashmaster:code(52997)]
+description=DRY DEP FLUX: ALICE (3D) PLEVS
+help=Dry deposition flux of ALICE
+ = on Pressure Levels
+
+[stashmaster:code(52998)]
+description=WET DEP FLUX: BOB (3D) PLEVS
+help=Wet deposition flux of BOB
+ = on Pressure Levels
+
[stashmaster:code(53181)]
description=Temperature Inc: Idealised (K/step)
help=Temperature increment from the idealised section in Kelvin per model
Index: rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster_A
===================================================================
--- rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster_A (revision 112818)
+++ rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster_A (revision 112862)
@@ -28373,6 +28373,24 @@
4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
5| 0 | 1871 | 0 | 65 | 0 | 0 | 0 | 0 | 0 |
#
+1| 1 | 50 | 996 |RXN FLUX: ALICE+OH->BOB(+Sec_Org) |
+2| 0 | 0 | 17 | 1 | 2 | 10 | 11 | 0 | 0 | 0 | 0 |
+3| 000000000000000000010000010000 | 00000000000000000001 | 3 |
+4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
+5| 0 | 0 | 0 | 65 | 0 | 0 | 0 | 0 | 0 |
+#
+1| 1 | 50 | 997 |DRY DEP FLUX: ALICE (3D) |
+2| 0 | 0 | 17 | 1 | 2 | 10 | 11 | 0 | 0 | 0 | 0 |
+3| 000000000000000000010000010000 | 00000000000000000001 | 3 |
+4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
+5| 0 | 0 | 0 | 65 | 0 | 0 | 0 | 0 | 0 |
+#
+1| 1 | 50 | 998 |WET DEP FLUX: BOB (3D) |
+2| 0 | 0 | 17 | 1 | 2 | 10 | 11 | 0 | 0 | 0 | 0 |
+3| 000000000000000000010000010000 | 00000000000000000001 | 3 |
+4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
+5| 0 | 0 | 0 | 65 | 0 | 0 | 0 | 0 | 0 |
+#
#===============================================================================
# Section 50 Item 999 reserved.
#===============================================================================
@@ -31658,6 +31676,24 @@
4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
5| 0 | 1871 | 0 | 8 | 0 | 0 | 0 | 0 | 0 |
#
+1| 1 | 52 | 996 |RXN FLUX: ALICE+OH->BOB PLEVS |
+2| 0 | 0 | 17 | 1 | 3 | 1 | 2 | 0 | 0 | 0 | 1 |
+3| 000000000000000000010000010000 | 00000000000000000001 | 3 |
+4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
+5| 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 |
+#
+1| 1 | 52 | 997 |DRY DEP FLUX: ALICE (3D) PLEVS |
+2| 0 | 0 | 17 | 1 | 3 | 1 | 2 | 0 | 0 | 0 | 1 |
+3| 000000000000000000010000010000 | 00000000000000000001 | 3 |
+4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
+5| 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 |
+#
+1| 1 | 52 | 998 |WET DEP FLUX: BOB (3D) PLEVS |
+2| 0 | 0 | 17 | 1 | 3 | 1 | 2 | 0 | 0 | 0 | 1 |
+3| 000000000000000000010000010000 | 00000000000000000001 | 3 |
+4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
+5| 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 |
+#
#===============================================================================
# Section 52 Item 999 reserved.
#===============================================================================
These differences can be found here:
Tutorials/UMvn13.0/worked_solutions/Task11.1/Task11.1_um.patch
The specific UKCA changes made are:
Index: src/control/core/diagnostics/asad_flux_dat.F90
===================================================================
--- src/control/core/diagnostics/asad_flux_dat.F90 (revision 568)
+++ src/control/core/diagnostics/asad_flux_dat.F90 (revision 570)
@@ -8424,6 +8424,32 @@
[' ',' ',' ',' ']) &
]
+! Additional diagnostics for UKCA tutorials
+! ST & CS chemistry version 131+ only
+! include Sec_Org if using aerosol chemistry
+TYPE(asad_flux_defn), PARAMETER, PUBLIC :: ukca_tutorial_fluxes_achem(3) = [ &
+asad_flux_defn('RXN',50996,'B',.FALSE.,0,4, &
+['ALICE ','OH '], &
+['BOB ','Sec_Org ',' ',' ']), &
+asad_flux_defn('DEP',50997,'D',.FALSE.,0,1, &
+['ALICE ',' '], &
+[' ',' ',' ',' ']), &
+asad_flux_defn('DEP',50998,'W',.FALSE.,0,1, &
+['BOB ',' '], &
+[' ',' ',' ',' ']) &
+]
+! don't include Sec_Org when not using aerosol chemistry
+TYPE(asad_flux_defn), PARAMETER, PUBLIC :: ukca_tutorial_fluxes(3) = [ &
+asad_flux_defn('RXN',50996,'B',.FALSE.,0,4, &
+['ALICE ','OH '], &
+['BOB ',' ',' ',' ']), &
+asad_flux_defn('DEP',50997,'D',.FALSE.,0,1, &
+['ALICE ',' '], &
+[' ',' ',' ',' ']), &
+asad_flux_defn('DEP',50998,'W',.FALSE.,0,1, &
+['BOB ',' '], &
+[' ',' ',' ',' ']) &
+]
PUBLIC :: asad_load_default_fluxes
@@ -8479,6 +8505,7 @@
INTEGER, PARAMETER :: ichem_version_117 = 117
INTEGER, PARAMETER :: ichem_version_119 = 119
INTEGER, PARAMETER :: ichem_version_121 = 121
+INTEGER, PARAMETER :: ichem_version_131 = 131
INTEGER :: i, j ! counters
! Generic temporary allocatable arrays
@@ -8509,6 +8536,7 @@
TYPE(asad_flux_defn), ALLOCATABLE, SAVE :: aa_ch4_budget_loss(:)
TYPE(asad_flux_defn), ALLOCATABLE, SAVE :: aa_ch4_drydep(:)
TYPE(asad_flux_defn), ALLOCATABLE, SAVE :: aa_ch4_ste(:)
+TYPE(asad_flux_defn), ALLOCATABLE, SAVE :: aa_ukca_tutorial_fluxes(:)
INTEGER :: p1 ! start position in asad_chemical_fluxes array
INTEGER :: p2 ! end position in asad_chemical_fluxes array
@@ -8658,6 +8686,16 @@
ALLOCATE(aa_ch4_ste(SIZE(asad_ch4_ste)))
aa_ch4_ste = asad_ch4_ste
END IF
+ ! aa_ukca_tutorial_fluxes
+ IF (ukca_config%i_ukca_chem_version >= ichem_version_131) THEN
+ IF (ukca_config%l_ukca_chem_aero) THEN
+ ALLOCATE(aa_ukca_tutorial_fluxes(SIZE(ukca_tutorial_fluxes_achem)))
+ aa_ukca_tutorial_fluxes = ukca_tutorial_fluxes_achem
+ ELSE
+ ALLOCATE(aa_ukca_tutorial_fluxes(SIZE(ukca_tutorial_fluxes)))
+ aa_ukca_tutorial_fluxes = ukca_tutorial_fluxes
+ END IF
+ END IF
ELSE IF (ukca_config%l_ukca_cristrat) THEN
! Select the asad diagnostics appropriate for CRI-Strat chemistry and
@@ -8866,6 +8904,16 @@
ALLOCATE(aa_oxidN_wetdep(SIZE(cri_oxidN_wetdep)))
aa_oxidN_wetdep = cri_oxidN_wetdep
END IF
+ ! aa_ukca_tutorial_fluxes
+ IF (ukca_config%i_ukca_chem_version >= ichem_version_131) THEN
+ IF (ukca_config%l_ukca_chem_aero) THEN
+ ALLOCATE(aa_ukca_tutorial_fluxes(SIZE(ukca_tutorial_fluxes_achem)))
+ aa_ukca_tutorial_fluxes = ukca_tutorial_fluxes_achem
+ ELSE
+ ALLOCATE(aa_ukca_tutorial_fluxes(SIZE(ukca_tutorial_fluxes)))
+ aa_ukca_tutorial_fluxes = ukca_tutorial_fluxes
+ END IF
+ END IF
ELSE IF (ukca_config%l_ukca_offline .OR. ukca_config%l_ukca_offline_be) THEN
@@ -8939,6 +8987,8 @@
n_chemical_fluxes = n_chemical_fluxes + SIZE(aa_ch4_drydep)
IF (ALLOCATED(aa_ch4_ste)) &
n_chemical_fluxes = n_chemical_fluxes + SIZE(aa_ch4_ste)
+IF (ALLOCATED(aa_ukca_tutorial_fluxes)) &
+ n_chemical_fluxes = n_chemical_fluxes + SIZE(aa_ukca_tutorial_fluxes)
ALLOCATE(asad_chemical_fluxes(n_chemical_fluxes))
@@ -9082,6 +9132,11 @@
asad_chemical_fluxes(p1:p2) = aa_ch4_ste(:)
p1 = p2 + 1
END IF
+IF (ALLOCATED(aa_ukca_tutorial_fluxes)) THEN
+ p2 = p1 + SIZE(aa_ukca_tutorial_fluxes) - 1
+ asad_chemical_fluxes(p1:p2) = aa_ukca_tutorial_fluxes(:)
+ p1 = p2 + 1
+END IF
IF (p2 /= n_chemical_fluxes) THEN
cmessage = ' n_chemical_fluxes and p2 are different'
@@ -9135,6 +9190,7 @@
END IF
! Deallocate the generic arrays
+IF (ALLOCATED(aa_ukca_tutorial_fluxes)) DEALLOCATE(aa_ukca_tutorial_fluxes)
IF (ALLOCATED(aa_ch4_ste)) DEALLOCATE(aa_ch4_ste)
IF (ALLOCATED(aa_ch4_drydep)) DEALLOCATE(aa_ch4_drydep)
IF (ALLOCATED(aa_ch4_budget_loss)) DEALLOCATE(aa_ch4_budget_loss)
These differences can be found here:
Tutorials/UMvn13.0/worked_solutions/Task11.1/Task11.1_ukca.patch
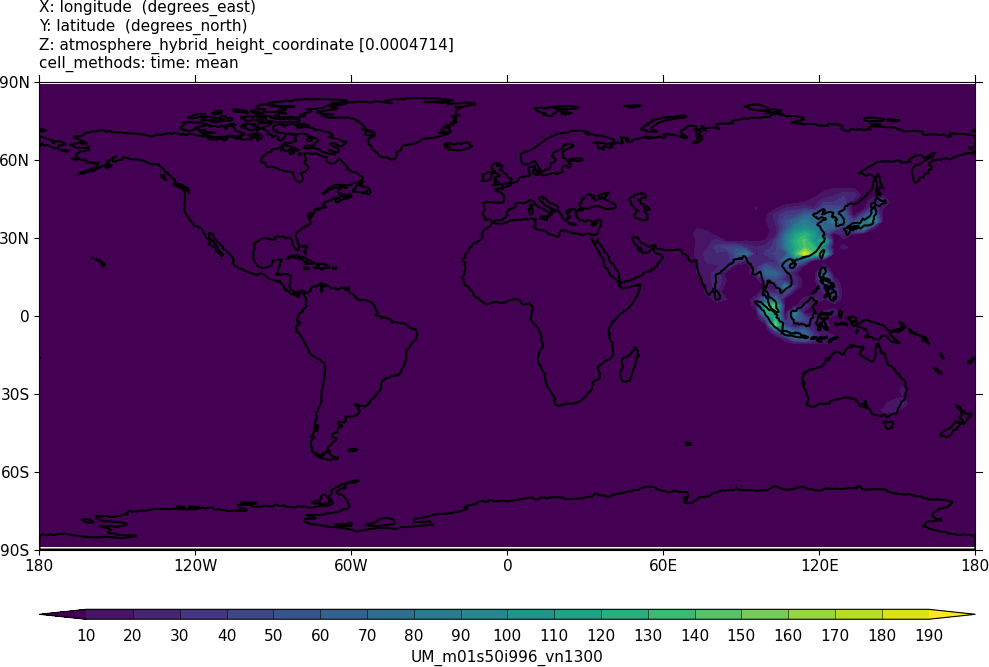
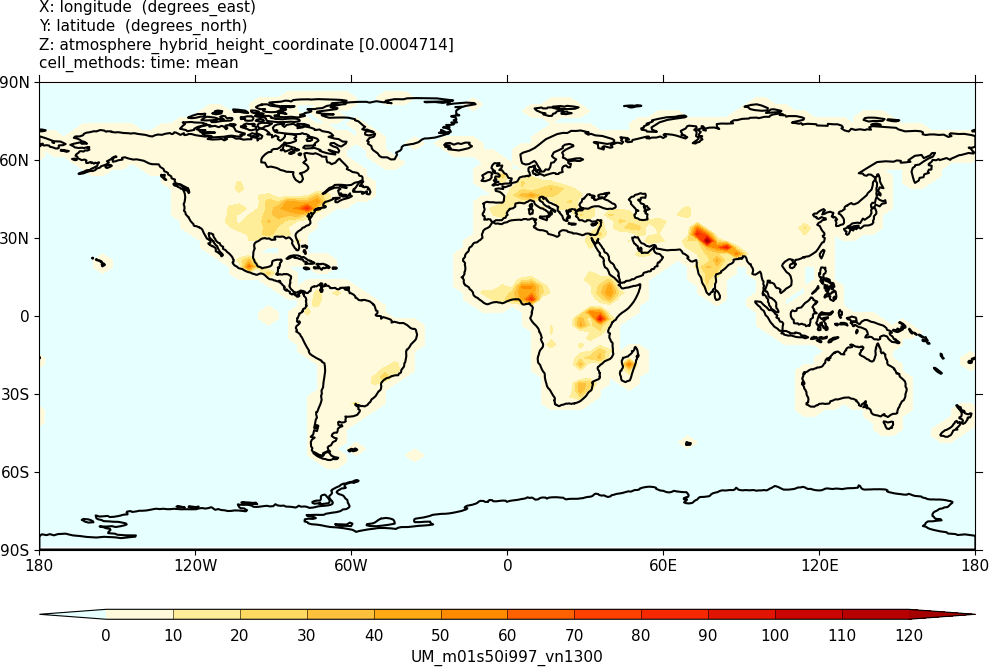
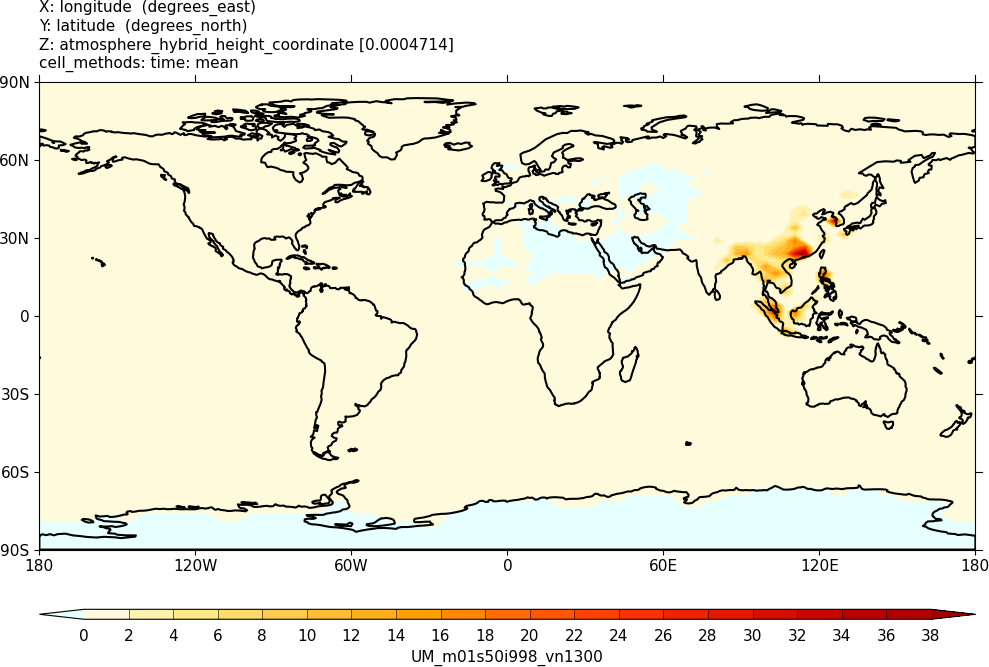
Sample output from this task can be found at
Tutorials/UMvn13.0/sample_output/Task11.1/
Checklist
- ☐ Make the required changes to branch's STASHmaster_A file, to add the new diagnostics. Make a note of the STASH items chosen, and also add the pressure-level diagnostics.
- ☐ Add help text for your diagnostics in your branch's STASHmaster-meta.conf file.
- ☐ Using a text editor, open the app/um/rose-app.conf file from your roses/[SUITE-ID] directory, and add the line STASHMASTER=STASHmaster in the [env] block, then save and close the file.
- ☐ Using a text editor, open the rose-suite.conf file from your roses/[SUITE-ID] directory, and add the following lines to the top of the file, before saving and closing it:
[file:app/um/file/STASHmaster] source=fcm:um.xm_br/dev/[your MOSRS userid]/vnX.Y_your_branch_name/rose-meta/um-atmos/vn11.8/etc/stash/STASHmaster@HEAD
- ☐ Point the metadata in your suite to the rose-meta/um-atmos/HEAD of your branch's working copy.
- ☐ Include your branch in your suite at: fcm_make env Sources.
- ☐ In asad_flux_dat.F90, make a new array of type asad_flux_defn and populate it with your new diagnostic specification(s), referencing the same item numbers as in your STASHmaster_A file.
- ☐ Append this new array at the end of asad_chemical_fluxes, and increment n_chemical_fluxes by the number of new diagnostics.
- ☐ If required, add new code for new diagnostics into asad_chem_flux_diags.F90 and other UKCA routines as necessary.
- ☐ Output your diagnostics in STASH at: um namelist Model Input and Output STASH Requests and Profiles STASH Requests.
- ☐ Run the TidyStashTransform transform macro.
- ☐ Save your suite.
- ☐ In the roses/[SUITE-ID] directory, run fcm commit to commit your changes to the repository.
- ☐ Run your suite.
UKCA Chemistry and Aerosol Tutorials at UMvn13.0
Written by Luke Abraham 2022