Difference between revisions of "Release Job UM11.0"
| Line 3: | Line 3: | ||
{| border="1" style="background-color:white" cellpadding="10" |
{| border="1" style="background-color:white" cellpadding="10" |
||
|- |
|- |
||
| − | || || XCE/F/S: '''<code>u-ay770</code>''' || Monsoon2: '''<code>u-ba055</code>''' |
+ | || || XCE/F/S: '''<code>u-ay770</code>''' || Monsoon2: '''<code>u-ba055</code>''' || NEXCS: '''<code></code>''' || ARCHER: '''<code>u-ba915</code>''' |
|- |
|- |
||
| − | || Run-time (per model month) || ~1 hour 10 minutes (XCS) || ~1 hour 10 minutes || ~1 hour 20 minutes |
+ | || Run-time (per model month) || ~1 hour 10 minutes (XCS) || ~1 hour 10 minutes || ~1 hour 10 minutes || ~1 hour 20 minutes |
|- |
|- |
||
| − | || Cycling || P1M in PT1H40M (XCS) || P1M in PT1H40M <br/> This could be changed to take advantage of the longer queues on NEXCS. || P1M in PT1H40M <br/> This could be changed to take advantage of the longer queues on ARCHER. |
+ | || Cycling || P1M in PT1H40M (XCS) || P1M in PT1H40M || P1M in PT1H40M <br/> This could be changed to take advantage of the longer queues on NEXCS. || P1M in PT1H40M <br/> This could be changed to take advantage of the longer queues on ARCHER. |
|- |
|- |
||
| − | || Estimated cost || || 168 node-hours/model year || |
+ | || Estimated cost || || 168 node-hours/model year <br/> or 91 kAU/model year equivalent || 168 node-hours/model year || 91 kAU/model year |
|- |
|- |
||
| − | || Decomposition || 18EW x 24NS on 12 nodes (XCS: 36PPN) || 18EW x 24NS on 12 nodes (36PPN) || 24EW x 15NS on 15 nodes (24PPN) |
+ | || Decomposition || 18EW x 24NS on 12 nodes (XCS: 36PPN) || 18EW x 24NS on 12 nodes (36PPN) || 18EW x 24NS on 12 nodes (36PPN) || 24EW x 15NS on 15 nodes (24PPN) |
|- |
|- |
||
| − | || Storage Requirements<br>(current STASH settings) || 73.5GB per model year (32-bit pp-files & seasonal 64-bit dumps) || 73.5GB per model year (32-bit pp-files & seasonal 64-bit dumps)<br>MASS costs: £2.01 per model year || 73.5GB per model year (32-bit pp-files & seasonal 64-bit dumps) <br/>copied to the <code>/nerc</code> disk and a JASMIN GWS as the model runs |
+ | || Storage Requirements<br>(current STASH settings) || 73.5GB per model year (32-bit pp-files & seasonal 64-bit dumps) || 73.5GB per model year (32-bit pp-files & seasonal 64-bit dumps)<br>MASS costs: £2.01 per model year || 73.5GB per model year (32-bit pp-files & seasonal 64-bit dumps) <br/>copied to the <code>/nerc</code> disk and a JASMIN GWS as the model runs || 73.5GB per model year (32-bit pp-files & seasonal 64-bit dumps) <br/>copied to the <code>/nerc</code> disk and a JASMIN GWS as the model runs |
|- |
|- |
||
|} |
|} |
||
Revision as of 14:55, 12 September 2018
Overview
XCE/F/S: u-ay770 |
Monsoon2: u-ba055 |
NEXCS: |
ARCHER: u-ba915
| |
| Run-time (per model month) | ~1 hour 10 minutes (XCS) | ~1 hour 10 minutes | ~1 hour 10 minutes | ~1 hour 20 minutes |
| Cycling | P1M in PT1H40M (XCS) | P1M in PT1H40M | P1M in PT1H40M This could be changed to take advantage of the longer queues on NEXCS. |
P1M in PT1H40M This could be changed to take advantage of the longer queues on ARCHER. |
| Estimated cost | 168 node-hours/model year or 91 kAU/model year equivalent |
168 node-hours/model year | 91 kAU/model year | |
| Decomposition | 18EW x 24NS on 12 nodes (XCS: 36PPN) | 18EW x 24NS on 12 nodes (36PPN) | 18EW x 24NS on 12 nodes (36PPN) | 24EW x 15NS on 15 nodes (24PPN) |
| Storage Requirements (current STASH settings) |
73.5GB per model year (32-bit pp-files & seasonal 64-bit dumps) | 73.5GB per model year (32-bit pp-files & seasonal 64-bit dumps) MASS costs: £2.01 per model year |
73.5GB per model year (32-bit pp-files & seasonal 64-bit dumps) copied to the /nerc disk and a JASMIN GWS as the model runs |
73.5GB per model year (32-bit pp-files & seasonal 64-bit dumps) copied to the /nerc disk and a JASMIN GWS as the model runs
|
UKCA Release Job - UM11.0 - Free Running
UM11.0 GA7.1 + UKCA Strattrop GLOMAP (N96L85) TS2000
Suite -id
Remember to always COPY these suites to a new one that you own, rather than checking them out. This will prevent you accidentally over-writing the settings in the original suite.
Base/ inherited configuration
UM11.0 GA7.1 UKCA Strattrop GLOMAP Y2014; TropO3 fixes suite u-av818 combined with settings from u-au917
UKCA evaluation suite output for u-au917 can be seen here
Branches
| Branch name | Description | Metadata change? |
|---|---|---|
| branches/dev/lukeabraham/vn11.0_ukca_linox_tweaks | Uses single NOx production factor for cld-cld and cldgrn flashes | Yes |
| branches/dev/dev/mohitdalvi/vn11.0_ukca_ageair_and_stashm | Fixes STASH definition of age-of-air pressure-level diagnostic Not required for build (fcm_make_um), only replaces runtime STASHmaster (specified in app/um/rose-app.conf=>[file:STASHmaster] section) |
Yes |
- Suite uses metadata from ~hadzm/code/vn11.0_ukca_linox_tweaks_meta/rose-meta/um-atmos/HEAD (same location on meto-cray and Monsoon2), as specified in app/um/rose-app.conf, meta=
Initialisation
The suite (meteorological, aerochem variables) will initialise from a dump produced at the end of a 20-year spin-up run of the previous GA7.1+Strattrop AMIP run u-au917 - /data/d05/hadzm/dumps/au917a.da20080901_00
Monsoon:/projects/ukca-meto/hadzm/dumps_ukmo/au9170a.da20080901_00
The path to this is set in the file(s): u-ay770/site/meto_cray.rc,u-ay770/site/MONSOON.rc as variable AINITIAL.
Forcing data
Emissions
Emissions for aerosol and chemical species (except biogenic) are prescribed as monthly cyclic climatologies around Year2000 by averaging data for 1995-2004 from the CMIP6 historical emissions developed for UKESM1. Script used was [attachment:make_clim_emiss.py]br
Biogenic emissions used are those originally developed for the NERC ACSIS project and represent datasets pertaining to 1990s,2000s for different species.
Sea surface temperature and sea ice concentrations
The SST and Sice values are also prescribed as monthly climatologies for 2000 created by averaging over 1995-2004 the timeseries data generated for CMIP6 Atmos-only simulations.
GHG and Trace Gas Concentrations
- These have been specified as values averaged over 1995-2004 from the historical GHG forcing data prepared for CMIP6.
- This was done by manually averaging data for above years from the files {{{/projects/ancils/cmip6/ancils/any/timeseries_1850-2014/cmip6/global_ghg_cmip6.conf}}} and {{{trgas_rcp_historical.dat}}} respectively for the Radiation GHG inputs and UKCA trace gas specification.
Critical Model setup / Namelist settings
SST/ Seaice
- Panel: um => namelist => Reconfiguration and Ancil control => Configure ancil and dump fields
- Look for initialisation of items:
stashcode=24 (SST), ancilfilename='$SST_SICE_ANCIL/sst_clim_mon_2000_n96e.anc' stashcodes=31,32 (sea-ice), ancilfilename='$SST_SICE_ANCIL/sice_clim_mon_2000_n96e.anc' site/meto_cray.rc: SST_SICE_CLIM = '/data/d05/hadzm/ancil/n96e/cmip6/sstice/clim_2000' site/MONSOON.rc: SST_SICE_CLIM = '/projects/ukca-meto/hadzm/ancil/n96e/cmip6/sstice/clim_2000'
RUN_Radiation GHG values
- Panel: Sec 01 02 Radiation => Gas MMRs => Varying Gas MMRs: Set l_climcfg=False and then specify these values in the Gas MMRs' panel:
| != Item = | = Value = | = Remarks = |
| c113mmr | 0.0 | |
| c114mmr | 0.0 | |
| c11mmr | 0.0 | |
| c12mmr | 4.3674e-09 | |
| ch4mmr | 9.799e-04 | ignored, uses conc from UKCA |
| co2_mmr | 5.5815e-04 | |
| hcfc22mmr | 0.0 | |
| hfc125mmr | 0.0 | |
| hfc134ammr | 3.5211e-10 | |
| n2ommr | 4.7797e-04 | ignored, uses conc from UKCA |
| o2mmr | 0.2314 |
RUN_UKCA settings
| Item | Value | Remarks |
|---|---|---|
| biom_aer_ems_scaling | 2.0 | |
| chem_timestep | 3600 | |
| dir_strat_aer | '/home/d05/hadzm/job_files/SAD' | |
| fastjx_mode | 1 | |
| fastjx_numwl | 18 | |
| fastjx_prescutoff | 20.0 | |
| file_strat_aer | 'CMIP_1850_2014_average_sad_v3_56_level.asc' | |
| i_ageair_reset_method | 1 | |
| i_mode_nzts | 15 | |
| i_mode_setup | 2 | |
| i_ukca_chem | 51 | |
| i_ukca_conserve_method | 2 | |
| i_ukca_dms_flux | 1 | |
| i_ukca_hiorder_scheme | 0 | |
| i_ukca_photol | 3 | |
| i_ukca_quasinewton_end | 3 | |
| i_ukca_quasinewton_start | 2 | |
| i_ukca_scenario | 2 | |
| i_ukca_solcyc | 0 | |
| jvscat_file | 'FJX_scat.dat' | |
| jvspec_dir | '$UMDIR/vn$VN/ctldata/UKCA/fastj' | |
| jvspec_file | 'FJX_spec_Nov11.dat' | |
| l_bcoc_bf | .true. | |
| l_bcoc_bm | .true. | |
| l_bcoc_ff | .true. | |
| l_mode_bhn_on | .true. | |
| l_mode_bln_on | .false. | |
| l_ukca | .true. | |
| l_ukca_ageair | .true. | |
| l_ukca_aie1 | .true. | |
| l_ukca_aie2 | .true. | |
| l_ukca_arg_act | .true. | |
| l_ukca_asad_columns | .false. | |
| l_ukca_asad_plev | .true. | |
| l_ukca_chem_aero | .true. | |
| l_ukca_chem_plev | .true. | |
| l_ukca_ddep_lev1 | .false. | |
| l_ukca_h2o_feedback | .true. | |
| l_ukca_het_psc | .true. | |
| l_ukca_ibvoc | .false. | |
| l_ukca_intdd | .true. | |
| l_ukca_limit_nat | .true. | |
| l_ukca_linox_scaling | .true. | |
| l_ukca_mode | .true. | |
| l_ukca_prescribech4 | .false. | |
| l_ukca_prim_moc | .true. | |
| l_ukca_primbcoc | .true. | |
| l_ukca_primss | .true. | |
| l_ukca_primsu | .true. | |
| l_ukca_qch4inter | .false. | |
| l_ukca_quasinewton | .true. | |
| l_ukca_radaer | .true. | |
| l_ukca_radaer_sustrat | .true. | |
| l_ukca_radch4 | .true. | |
| l_ukca_radf11 | .false. | |
| l_ukca_radf113 | .false. | |
| l_ukca_radf12 | .false. | |
| l_ukca_radf22 | .false. | |
| l_ukca_radn2o | .true. | |
| l_ukca_rado3 | .true. | |
| l_ukca_sa_clim | .true. | |
| l_ukca_scale_biom_aer_ems | .true. | |
| l_ukca_scale_seadms_ems | .false. | |
| l_ukca_scale_soa_yield | .true. | |
| l_ukca_set_trace_gases | .true. | |
| l_ukca_sfix | .false. | |
| l_ukca_so2ems_expvolc | .false. | |
| l_ukca_src_in_conservation | .true. | |
| l_ukca_trophet | .false. | |
| l_ukca_use_background_aerosol | .true. | |
| lightnox_scale_fac | 2.0 | |
| max_ageair_reset_level | 10 | |
| mode_activation_dryr | 37.5 | |
| mode_aitsol_cvscav | 0.5 | |
| mode_incld_so2_rfrac | 0.25 | |
| mode_parfrac | 2.5 | |
| soa_yield_scaling | 2.0 | |
| ukca_em_dir | Top-level paths: CMIP6_AEROCHEM_EMS = /data/d05/hadzm/ancil/n96e/cmip6/ !AeroChemEms/clim_2000/v1 (Meto-Cray) = /projects/ukca-meto/hadzm/ancil/n96e/cmip6/ !AeroChemEms/clim_2000/v1 (Monsoon2) CMIP6_BIOG_EMS = /projects/um1/ancil/atmos/ n96e/ukca_emiss/biogenic/v1 (Both systems). Locations are set in file <suite>/site/<site>.rc | |
| ukca_em_files | '$CMIP6_AEROCHEM_EMS/BC_biofuel_anthropogenic_1995_2004_clim.nc' '$CMIP6_AEROCHEM_EMS/BC_fossil_anthropogenic_1995_2004_clim.nc' '$CMIP6_AEROCHEM_EMS/DMS_biomass_low_1995_2004_clim.nc' '$CMIP6_BIOG_EMS/DMS_land_spiro1992.nc' '$CMIP6_AEROCHEM_EMS/OC_biofuel_anthropogenic_1995_2004_clim.nc' '$CMIP6_AEROCHEM_EMS/OC_fossil_anthropogenic_1995_2004_clim.nc' '$CMIP6_AEROCHEM_EMS/SO2_high_anthropogenic_1995_2004_clim.nc' '$CMIP6_AEROCHEM_EMS/SO2_low_anthropogenic_1995_2004_clim.nc' '$UM_NETCDF_UKCAEMISS_SO2NAT_DIR/$UM_NETCDF_UKCAEMISS_SO2NAT_FILE' '$CMIP6_AEROCHEM_EMS/BC_biomass_high_1995_2004_clim.nc' '$CMIP6_AEROCHEM_EMS/BC_biomass_low_1995_2004_clim.nc' '$CMIP6_AEROCHEM_EMS/OC_biomass_high_1995_2004_clim.nc' '$CMIP6_AEROCHEM_EMS/OC_biomass_low_1995_2004_clim.nc' '$CMIP6_BIOG_EMS/MEGAN-MACC_biogenic_C2H6_clim_2001-2010.nc' | |
| ukca_h2mmr | 3.4528E-08 | |
| ukca_n2mmr | 7.5468E-01 | |
| ukca_offline_dir | ignored | |
| ukca_offline_files | ignored | |
| ukca_rcpdir | '/home/d05/hadzm/job_files/scenario_files' | |
| ukca_rcpfile | 'trgas_rcp_clim_2000.dat' | |
| ukcaaclw | '$UMDIR/vn$VN/ctldata/UKCA/radaer/ga7_1/nml_ac_lw' | |
| ukcaacsw | '$UMDIR/vn$VN/ctldata/UKCA/radaer/ga7_1/nml_ac_sw' | |
| ukcaanlw | '$UMDIR/vn$VN/ctldata/UKCA/radaer/ga7_1/nml_an_lw' | |
| ukcaansw | '$UMDIR/vn$VN/ctldata/UKCA/radaer/ga7_1/nml_an_sw' | |
| ukcacrlw | '$UMDIR/vn$VN/ctldata/UKCA/radaer/ga7_1/nml_cr_lw' | |
| ukcacrsw | '$UMDIR/vn$VN/ctldata/UKCA/radaer/ga7_1/nml_cr_sw' | |
| ukcaprec | '$SPECTRAL_FILE_DIR/RADAER_pcalc.ukca' |
STASH requests
Diagnostics Requested in u-ay770 can be seen here
Assessment
UKCA evaluation suite output for : ![]() Chemistry
Chemistry ![]() and
and ![]() Aerosols
Aerosols ![]()
Autoassess output (vs UM11.0 GA7): Internal, External. Note: Radiation area output not produced. Click on link for Aerosol area output.
Validation notes: Internal, External
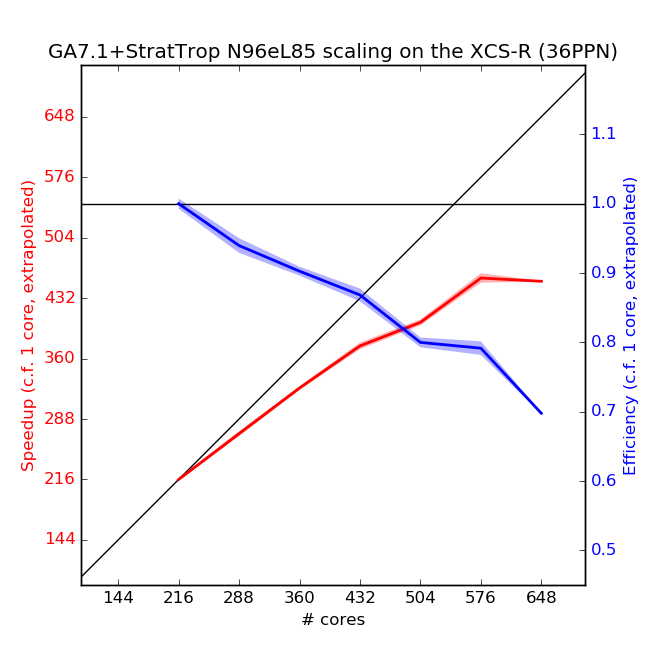
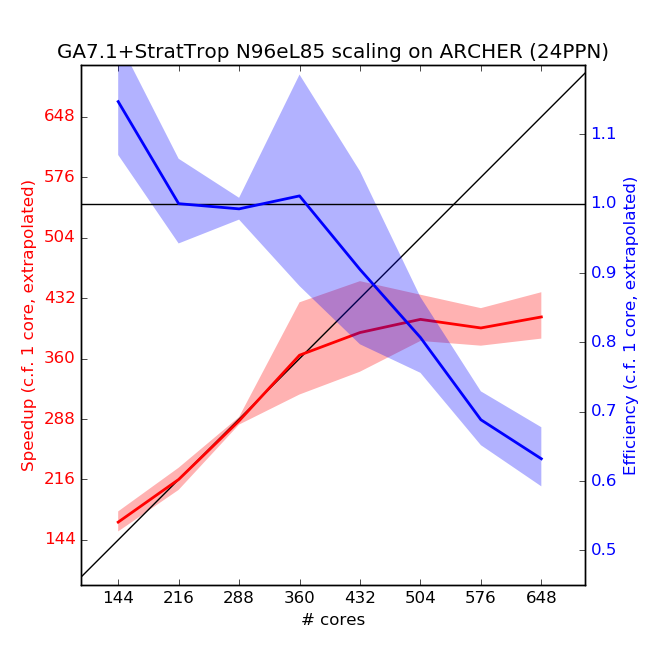
Scaling Tests
Scaling tests of GA7.1+StratTrop have been performed on the XCS and ARCHER using UM vn10.6.1. Scaling plots are below.
Monsoon2 changes
You will likely need to correctly set your Monsoon2 project and MOOSE project name in the suite. To do this go to suite conf Host Machine Met Office / Monsoon. You should then:
- Set Account_Monsoon to the correct value, e.g. ukca-cam etc.
- Put the correct MOOSE project name if required, e.g. ukca. Your Monsoon2 MOOSE project will be of the form project-NAME, and you only need to enter NAME here.
ARCHER archiving to the RDF and JASMIN
The ARCHER suite has been configured to allow for automatic archiving to the RDF (/nerc disk) and a JASMIN Group Workspace if required. These settings are found in the postproc Post Processing and then either select Archer Archiving or JASMIN Transfer.
Archer Archiving using the postproc task
- Set the value of archive_root_path to be on the
/nercdisk, e.g./nerc/n02/n02/Auserid/archive, whereAuseridis your ARCHER userid. - Turn on archiving via the postproc task, by clicking the suite conf Tasks Post Processing button.
JASMIN Transfer using the pptransfer task
- Ensure that you have archiving to the RDF turned on using the postproc task (see above).
- Set the value of transfer_dir to be the path to a JASMIN Group Workspace, e.g.
/group_workspaces/jasmin2/ukca/vol1/Juserid/archive, whereJuseridis your JASMIN userid. You can apply for access to a GWS on the JASMIN website. - Set the value of remote_host to be the transfer machine on JASMIN to use, e.g.
jasmin-xfer1.ceda.ac.ukorjasmin-xfer2.ceda.ac.uk. Usingjasmin-xfer2is recommended but you must first request access to hpxfer. - You will need to ensure that you can connect from PUMA to the RDF data transfer node (e.g.
dtn02.rdf.ac.uk). - You will need to ensure that you can connect from the data transfer node to JASMIN.
- You will need to ensure you include the following in your RDF
~/.ssh/configfile:
Host jasmin-xfer2 jasmin-xfer2.ceda.ac.uk
Hostname jasmin-xfer2.ceda.ac.uk
User Juserid
IdentityFile ~/.ssh/id_rsa_jasmin
ForwardAgent no
Host jasmin-xfer1 jasmin-xfer1.ceda.ac.uk
Hostname jasmin-xfer1.ceda.ac.uk
User Juserid
IdentityFile ~/.ssh/id_rsa_jasmin
ForwardAgent no
- where
Juseridis your JASMIN userid. You should replace~/.ssh/id_rsa_jasminwith the path to your JASMIN SSH key (if it is named differently).
- You will need to ensure that you have the following in your PUMA
~/.ssh/configfile:
Host dtn02 dtn02.rdf.ac.uk
Hostname dtn02.rdf.ac.uk
User Auserid
IdentityFile ~/.ssh/id_dsa
ForwardAgent no
- where
Auseridis your ARCHER/RDF userid. You should replace~/.ssh/id_dsawith the path to your ARCHER SSH key (if it is named differently).
- Turn on the pptransfer task by clicking the suite conf Tasks PP Transfer button.