Difference between revisions of "UKCA Chemistry and Aerosol vn11.8 Tutorial 4"
| (32 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[UKCA Chemistry and Aerosol Tutorials at vn11.8]] |
[[UKCA Chemistry and Aerosol Tutorials at vn11.8]] |
||
| + | |||
| + | {| class="wikitable" border = "1px" |
||
| + | |- |
||
| + | | Difficulty || '''<span style="color:red">HARD</span>''' |
||
| + | |- |
||
| + | | Time to Complete || '''2 or more hours''' |
||
| + | |- |
||
| + | | Video instructions || '''https://youtu.be/6XqYfW2qHKY?t=213''' |
||
| + | |} |
||
| + | |||
| + | '''Remember to run <tt>mosrs-cache-password</tt> in your Rose suite and UM branch terminals.''' |
||
==What you will do in this tutorial== |
==What you will do in this tutorial== |
||
| Line 8: | Line 19: | ||
<span style="color:green">'''TASK4.1:''' Add in two new tracers in to slots 498 and 499 for the StratTrop chemistry scheme. The tracer in slot 498 will be called '''ALICE''' and the tracer in slot 499 will be called '''BOB'''. You should initialise these tracers to <code>1.000000e-15</code>. You should also output these two tracers through the '''UPA''' stream in STASH as 3-hour means. Do not make any UKCA code changes at this stage.</span> |
<span style="color:green">'''TASK4.1:''' Add in two new tracers in to slots 498 and 499 for the StratTrop chemistry scheme. The tracer in slot 498 will be called '''ALICE''' and the tracer in slot 499 will be called '''BOB'''. You should initialise these tracers to <code>1.000000e-15</code>. You should also output these two tracers through the '''UPA''' stream in STASH as 3-hour means. Do not make any UKCA code changes at this stage.</span> |
||
| + | |||
| + | '''Note:''' if you are working at UM versions prior to UMvn11.7, you will only be able to use tracer slots up to 256 (rather than 499 as used here). For more information see the [[UKCA_Chemistry_and_Aerosol_vn10.9_Tutorial_4|vn10.9 Tutorials]]. |
||
==Make a branch== |
==Make a branch== |
||
| Line 39: | Line 52: | ||
====Using your new metadata within Rose==== |
====Using your new metadata within Rose==== |
||
| + | Here you should click on the '''um''' app in Rose and change the '''meta''' setting from <code>um-atmos/vn11.8</code> to |
||
| − | There are two ways to ensure that your metadata is used within Rose: |
||
| − | |||
| − | ===== 1. Open suites using <code>-M</code> ===== |
||
| − | |||
| − | To pick-up the metadata changes, you should make sure that the '''meta''' path in '''um''' points to |
||
| − | |||
| − | um-atmos/HEAD |
||
| − | |||
| − | From now on, when you want to open your Rose suite for editing, instead of doing it through rosie, you should instead open the job for editing by going to your <code>$HOME/roses/'''[SUITE-ID]'''</code> directory, and opening it using the command |
||
| − | |||
| − | rose edit -M /path/to/vn11.8_<span style="color:blue">your_branch_name</span>/rose-meta |
||
| − | |||
| − | where here '''<code>/path/to</code>''' means the full path to your suite on the machine that you are using (e.g. PUMA, the VM, etc.). If you don't do this, the STASHmaster changes will not be picked up, and you won't be able to select the tracers. |
||
| − | |||
| − | ===== 2. Edit the <code>meta</code> path in your um app ===== |
||
| − | |||
| − | Here you should click on the '''um''' app in Rose and change the '''meta''' setting from <code>um-atmos/vn11.</code> to |
||
/path/to/vn11.8_<span style="color:blue">your_branch_name</span>/rose-meta/um-atmos/HEAD |
/path/to/vn11.8_<span style="color:blue">your_branch_name</span>/rose-meta/um-atmos/HEAD |
||
| Line 69: | Line 66: | ||
/path/to/vn11.8_<span style="color:blue">your_branch_name</span> |
/path/to/vn11.8_<span style="color:blue">your_branch_name</span> |
||
| − | if you wish to run from a working copy, and |
+ | (where <tt>/path/to/vn11.8_your_branch_name</tt> means the full linux path to your branch, e.g. <tt>/home/ukcatr01/vn11.8_UKCA_Tutorials</tt> etc.) if you wish to run from a working copy, and |
branches/dev/'''<span style="color:green">[your MOSRS userid]</span>'''/vn11.8_<span style="color:blue">your_branch_name</span> |
branches/dev/'''<span style="color:green">[your MOSRS userid]</span>'''/vn11.8_<span style="color:blue">your_branch_name</span> |
||
| Line 79: | Line 76: | ||
fcm info fcm:um.xm |
fcm info fcm:um.xm |
||
| − | and check the last committed revision. If it is the same or higher than your last revision then your code changes will be picked-up. If it is less than your revision your code changes will not be included in the compile. On the VM, this is not an issue as there is no mirror. |
+ | and check the last committed revision. If it is the same or higher than your last revision then your code changes will be picked-up. If it is less than your revision your code changes will not be included in the compile. '''On the VM, this is not an issue as there is no mirror.''' |
===STASHmaster_A=== |
===STASHmaster_A=== |
||
| Line 106: | Line 103: | ||
* '''<span style="color:red">34</span>:''' The ''STASH section'', in this case Section 34, UKCA prognostics. |
* '''<span style="color:red">34</span>:''' The ''STASH section'', in this case Section 34, UKCA prognostics. |
||
* '''<span style="color:red">1</span>:''' The ''STASH item'', in this case, the first item in s34, ozone. |
* '''<span style="color:red">1</span>:''' The ''STASH item'', in this case, the first item in s34, ozone. |
||
| − | * '''<span style="color:red">O3 MASS MIXING RATIO AFTER TIMESTEP</span>:''' The ''name'' of the field in the STASHmaster file. This can be anything descriptive, but it isn't |
+ | * '''<span style="color:red">O3 MASS MIXING RATIO AFTER TIMESTEP</span>:''' The ''name'' of the field in the STASHmaster file. This can be anything descriptive, but it isn't used by the model, and is only considered in Rose (or e.g. Xconv). |
* |
* |
||
| Line 116: | Line 113: | ||
'''Third Line:''' |
'''Third Line:''' |
||
| − | * '''<span style="color:red">000000000000000000100000111110</span>:''' These are ''Options Codes'' which determine which scheme the STASH field (i.e. tracer in this case) are valid for. These are counted from the '''right''', '''''n1''''' to '''''n30''''' |
+ | * '''<span style="color:red">000000000000000000100000111110</span>:''' These are ''Options Codes'' which determine which scheme the STASH field (i.e. tracer in this case) are valid for. These are counted from the '''right''', '''''n1''''' to '''''n30'''''. Only some of these are used, and have the following meaning: |
** '''n1:''' Age-of-air tracer (''ageair'') or aerosol tracers |
** '''n1:''' Age-of-air tracer (''ageair'') or aerosol tracers |
||
** '''n2:''' Standard Tropospheric Chemistry scheme using the Backward Euler solver (''trop'') |
** '''n2:''' Standard Tropospheric Chemistry scheme using the Backward Euler solver (''trop'') |
||
| Line 163: | Line 160: | ||
You will notice that there are a number of changes to various codes, and this is to do with defining the grid that the diagnostic is valid on, whether it can be held in the dump, etc. For more information on what these codes mean, please see Appendix C in UMDP C4, which can be obtained from the SRS [https://code.metoffice.gov.uk/doc/um/latest/umdp.html here (password required)]. When making a new entry, the easiest thing to do is copy an existing entry and make the required changes to the item, name, PP field code, and option codes. |
You will notice that there are a number of changes to various codes, and this is to do with defining the grid that the diagnostic is valid on, whether it can be held in the dump, etc. For more information on what these codes mean, please see Appendix C in UMDP C4, which can be obtained from the SRS [https://code.metoffice.gov.uk/doc/um/latest/umdp.html here (password required)]. When making a new entry, the easiest thing to do is copy an existing entry and make the required changes to the item, name, PP field code, and option codes. |
||
| − | <span style="color:green">'''Remember:''' You need to add '''2''' tracers, ALICE in s34i498 and BOB in s34i499.</span> |
+ | <span style="color:green">'''Remember:''' You need to add '''2''' tracers, ALICE in s34i498 and BOB in s34i499 (and section 51 for pressure level equivalents).</span> |
====Help text==== |
====Help text==== |
||
| Line 175: | Line 172: | ||
and |
and |
||
| − | [stashmaster:code( |
+ | [stashmaster:code(51498)] |
description=ALICE MASS MIX RATIO ON PRESS LEVS |
description=ALICE MASS MIX RATIO ON PRESS LEVS |
||
help=UKCA Tutiorial Tracer ALICE Mass Mixing Ratio in kg/kg(Air) |
help=UKCA Tutiorial Tracer ALICE Mass Mixing Ratio in kg/kg(Air) |
||
=on pressure levels |
=on pressure levels |
||
| − | <span style="color:green">'''Remember:''' You need to add '''2''' tracers, ALICE in s34i498 and BOB in s34i499.</span> |
+ | <span style="color:green">'''Remember:''' You need to add '''2''' tracers, ALICE in s34i498 and BOB in s34i499 (and section 51 for pressure level equivalents).</span> |
'''You should make these changes to STASHmaster_A and STASHmaster-meta.conf, and then <code>fcm commit</code> these changes. This is important, otherwise the running suite will not be able to see the changes you have made once you have included your new STASHmaster_A file in Rose.''' |
'''You should make these changes to STASHmaster_A and STASHmaster-meta.conf, and then <code>fcm commit</code> these changes. This is important, otherwise the running suite will not be able to see the changes you have made once you have included your new STASHmaster_A file in Rose.''' |
||
| Line 190: | Line 187: | ||
To get Rose to recognise your STASHmaster file, you will need to make a number of changes: |
To get Rose to recognise your STASHmaster file, you will need to make a number of changes: |
||
| − | # Make sure to point the '''um''' metadata to your branch (if it isn't already) by |
+ | # Make sure to point the '''um''' metadata to your branch (if it isn't already) by editing the '''um''' '''meta''' path (you should already be doing this after following the instructions in the [[#Metadata|metadata section]] above). |
# You need to open the <code>$HOME/roses/'''[SUITE-ID]'''/app/um/rose-app.conf</code> file in a text editor (e.g. vim, emacs, nedit etc.) and add the line <code>STASHMASTER=STASHmaster</code> inside the '''<code>[env]</code>''' block near the top of the file. |
# You need to open the <code>$HOME/roses/'''[SUITE-ID]'''/app/um/rose-app.conf</code> file in a text editor (e.g. vim, emacs, nedit etc.) and add the line <code>STASHMASTER=STASHmaster</code> inside the '''<code>[env]</code>''' block near the top of the file. |
||
# You need to open the <code>$HOME/roses/'''[SUITE-ID]'''/rose-suite.conf</code> file in a text editor and add the following lines at the top of the file: |
# You need to open the <code>$HOME/roses/'''[SUITE-ID]'''/rose-suite.conf</code> file in a text editor and add the following lines at the top of the file: |
||
| Line 236: | Line 233: | ||
/home/'''[your VM userid]'''/cylc-run/'''[SUITE-ID]'''/work/1/atmos/ |
/home/'''[your VM userid]'''/cylc-run/'''[SUITE-ID]'''/work/1/atmos/ |
||
| + | |||
| + | === Viewing output in Xconv === |
||
| + | |||
| + | If you open the '''.pa''' file in Xconv, you should now see the following if you have outputted both the section 34 and section 51 fields: |
||
| + | |||
| + | 4 : 96 72 27 1 Stash code = 51498 |
||
| + | 5 : 96 72 27 1 Stash code = 51499 |
||
| + | |||
| + | 11 : 96 72 38 1 Stash code = 34498 |
||
| + | 12 : 96 72 38 1 Stash code = 34499 |
||
| + | |||
| + | '''Note here that these fields are not named <tt>ALICE</tt> and <tt>BOB</tt>, or however you have described them in your STASHmaster_A file.''' They are instead labelled by the unique STASH code identifier that you have defined for them. This is working as expected. |
||
| + | |||
| + | * '''Optional:''' It is possible (although not necessary) to tell Xconv to read the STASHmaster_A file from your branch. To do this, in Xconv go to '''Setup <math>\rightarrow</math> Select STASH Master Files''' and then navigate to the location of the file in your branch's working copy in the left-hand side of the panel. When you have located it, highlight it and click '''Add'''. Then click '''Apply''', '''Save''', and '''Dismiss'''. Now Xconv will list the names of your species as you have called them in your STASHmaster_A file. |
||
===Numerical Noise=== |
===Numerical Noise=== |
||
| Line 244: | Line 255: | ||
</gallery> |
</gallery> |
||
| − | When your suite has run successfully, and you view the fields for your new tracer(s) in Xconv, you may find that they have a speckled appearance, rather than being constant. This is due to numerical noise that has been introduced as the tracers go through the UM timesteps. Once the field is converted to 32-bit this should disappear. It will also cease to become apparent when emissions and reactions are applied to these tracers. |
+ | When your suite has run successfully, and you view the fields for your new tracer(s) in Xconv, you may find that they have a speckled appearance, rather than being constant. This is due to numerical noise that has been introduced as the tracers go through the UM timesteps. Once the field is converted to a 32-bit pp-file this should disappear. It will also cease to become apparent when emissions and reactions are applied to these tracers. |
==Solution to Task4.1== |
==Solution to Task4.1== |
||
| Line 469: | Line 480: | ||
==Required UKCA Code Changes== |
==Required UKCA Code Changes== |
||
| + | |||
| + | Note that at some older UKCA versions it was necessary to make the UKCA changes in parallel with the UM-side changes covered above. |
||
===<code>ukca_nmspec_mod.F90</code>=== |
===<code>ukca_nmspec_mod.F90</code>=== |
||
| Line 583: | Line 596: | ||
When adding new tracers you should not overwrite tracers that are already in use by the chemistry scheme that you are using (e.g. StratTrop etc.), and it is best to also take care and avoid tracers in use by other schemes, if possible. You can see the tracers used by each scheme by looking in the <code>chch_defs</code> array at the top of the <code>ukca_chem_master.F90</code> module. |
When adding new tracers you should not overwrite tracers that are already in use by the chemistry scheme that you are using (e.g. StratTrop etc.), and it is best to also take care and avoid tracers in use by other schemes, if possible. You can see the tracers used by each scheme by looking in the <code>chch_defs</code> array at the top of the <code>ukca_chem_master.F90</code> module. |
||
| − | Here you should put a new species, <tt>'ALICE '</tt>, into tracer slot 498 (currently specified as < |
+ | Here you should put a new species, <tt>'ALICE '</tt>, into tracer slot 498 (currently specified as <tt>'XXX '</tt>, which means that it is empty and is not in use by any scheme at the moment). |
| − | <span style="color:green">'''Remember:''' You need to add '''2''' tracers, ALICE in s34i498 and BOB in |
+ | <span style="color:green">'''Remember:''' You need to add '''2''' tracers, ALICE in s34i498 and BOB in s34i499.</span> |
===<code>ukca_chem_master.F90</code>=== |
===<code>ukca_chem_master.F90</code>=== |
||
| − | We will go through changes that need to be made for the StratTrop chemistry scheme (defined in the file '''<code>ukca_chem_master.F90</code>''', using the code '''<code> |
+ | We will go through changes that need to be made for the StratTrop chemistry scheme (defined in the file '''<code>ukca_chem_master.F90</code>''', using the code '''<code>st</code>'''), but equivalent changes would need to be made for others, e.g. TropIsop or Strat. For this example we will only refer to this scheme. |
In this file the list of species used in chemistry is defined by the array '''<code>chch_defs_master</code>'''. The list of species is held in a ''derived type'', of the format: |
In this file the list of species used in chemistry is defined by the array '''<code>chch_defs_master</code>'''. The list of species is held in a ''derived type'', of the format: |
||
| Line 661: | Line 674: | ||
===<code>ukca_cspecies.F90</code>=== |
===<code>ukca_cspecies.F90</code>=== |
||
| − | After you have added in the '''C_<span style="color:blue">species</span>''' conversion factors, you will need to tell UKCA to use them for your species. To do this you need to edit the '''ukca_cspecies.F90''' module, which contains code which |
+ | After you have added in the '''C_<span style="color:blue">species</span>''' conversion factors, you will need to tell UKCA to use them for your species. To do this you need to edit the '''ukca_cspecies.F90''' module, which contains code which constructs the '''c_species''' array of conversion factors for the advected tracers. This contains a subroutine called <code>ukca_calc_cspecies</code> which has a long block of code that you need to edit to add an entry like this: |
WHERE (advt == 'ALICE ') c_species = C_ALICE |
WHERE (advt == 'ALICE ') c_species = C_ALICE |
||
The '''advt''' array is automatically generated by UKCA at run-time from the <code>chch_defs_master</code> chemistry scheme definition you edited earlier, so your new tracer(s) will exist within it. You need to add in a new line for each of your tracers which sets the value of the '''c_species''' array to your individual C_<span style="color:blue">species</span> parameter. Add the line(s) in at the end of the block. |
The '''advt''' array is automatically generated by UKCA at run-time from the <code>chch_defs_master</code> chemistry scheme definition you edited earlier, so your new tracer(s) will exist within it. You need to add in a new line for each of your tracers which sets the value of the '''c_species''' array to your individual C_<span style="color:blue">species</span> parameter. Add the line(s) in at the end of the block. |
||
| + | |||
| + | As the <tt>C_<span style="color:blue">species</span></tt> conversion factors are defined in the <code>ukca_constants</code> module you will need to explicitly add them to the |
||
| + | |||
| + | USE ukca_constants, ONLY: ... |
||
| + | |||
| + | statement at the top of the module. |
||
<span style="color:green">'''Remember:''' You need to add '''2''' tracers, ALICE in s34i498 and BOB in s34i499.</span> |
<span style="color:green">'''Remember:''' You need to add '''2''' tracers, ALICE in s34i498 and BOB in s34i499.</span> |
||
| Line 671: | Line 690: | ||
==Run your suite again== |
==Run your suite again== |
||
| − | Now that you have made the required changes to Rose and in your branch, please run your suite again. |
+ | Now that you have made the required changes to Rose and in your branch, please add your branch to your suite in Rose and run your suite again. To include the branch in your suite add it to the '''fcm_make <math>\rightarrow</math> env <math>\rightarrow</math> Sources''' panel. In the '''<code>um_sources</code>''' entry click the '''green cross (<span style="color:green">+</span>)''' and put your branch in the box like |
| + | |||
| + | branches/dev/'''<span style="color:green">[your MOSRS userid]</span>'''/vn11.8_<span style="color:blue">your_branch_name</span> |
||
| + | |||
| + | This will take the latest revision from the repository that you have checked-in (but '''not''' any code that you have not committed). You can also specify a fixed revision for a branch, e.g. |
||
| + | |||
| + | branches/dev/'''<span style="color:green">[your MOSRS userid]</span>'''/vn11.8_<span style="color:blue">your_branch_name</span>@<span style="color:red">123456</span> |
||
| + | |||
| + | '''In this case you will need to remember to update your branch revision number every time you commit a change.''' |
||
| + | |||
| + | You could also point to a working copy which will take the code as it is on the disk, including any code not committed to the repository, e.g. |
||
| + | |||
| + | /home/'''[your VM userid]'''/vn11.8_<span style="color:blue">your_branch_name</span> |
||
| + | |||
| + | and here you should '''not''' specify any revision numbers. |
||
| + | |||
| + | === Failures and Debugging === |
||
| + | |||
| + | You may find that it fails on the <code>fcm_make</code>, <code>recon</code>, or <code>atmos</code> jobs. To find the errors and see output, you should either right-click the failed task in Gcylc and view the output, or go to the |
||
/home/'''[your VM userid]'''/cylc-run/'''[SUITE-ID]''' |
/home/'''[your VM userid]'''/cylc-run/'''[SUITE-ID]''' |
||
| Line 682: | Line 719: | ||
/home/'''[your VM userid]'''/cylc-run/'''[SUITE-ID]'''/work/1/atmos/ |
/home/'''[your VM userid]'''/cylc-run/'''[SUITE-ID]'''/work/1/atmos/ |
||
| + | |||
| + | If you have errors in the '''fcm_make''' task it will give a line number in the Fortran file for where the error is. However, this line number will not be for your branch in your working copy, e.g. |
||
| + | |||
| + | /home/'''[your VM userid]'''/vn11.8_<span style="color:blue">your_branch_name</span> |
||
| + | |||
| + | but will instead be for code inside your <code>cylc-run</code> directory. |
||
| + | |||
| + | ==== Example ==== |
||
| + | |||
| + | If we consider the following error: |
||
| + | |||
| + | [FAIL] mpif90 -oo/ukca_nmspec_mod.o -c -I./include -I/home/vagrant/cylc-run/vn11.8_prebuilds/share/fcm_make_vm_gnu_um_safe_omp/build-atmos/include -fdefault-integer-8 -fdefault-real-8 -fdefault-double-8 -std=f2008ts -Wtabs -I/home/vagrant/umdir/gcom/default/vm_gfortran_mpich3/build/include -O2 -Werror -fopenmp -I/usr/include -I/usr/include /home/ubuntu/cylc-run/u-ca024/share/fcm_make/preprocess-atmos/src/um/src/atmosphere/UKCA/ukca_nmspec_mod.F90 # rc=1 |
||
| + | [FAIL] /home/ubuntu/cylc-run/u-ca024/share/fcm_make/preprocess-atmos/src/um/src/atmosphere/UKCA/ukca_nmspec_mod.F90:160:31: |
||
| + | [FAIL] |
||
| + | [FAIL] 'XXX ','XXX ',ALICE ,BOB & !499 |
||
| + | [FAIL] 1 |
||
| + | [FAIL] Error: Symbol \u2018alice\u2019 at (1) has no IMPLICIT type |
||
| + | [FAIL] /home/ubuntu/cylc-run/u-ca024/share/fcm_make/preprocess-atmos/src/um/src/atmosphere/UKCA/ukca_nmspec_mod.F90:160:36: |
||
| + | [FAIL] |
||
| + | [FAIL] 'XXX ','XXX ',ALICE ,BOB & !499 |
||
| + | [FAIL] 1 |
||
| + | [FAIL] Error: Symbol \u2018bob\u2019 at (1) has no IMPLICIT type |
||
| + | [FAIL] compile 2.2 ! ukca_nmspec_mod.o <- um/src/atmosphere/UKCA/ukca_nmspec_mod.F90 |
||
| + | [FAIL] ! ukca_nmspec_mod.mod : depends on failed target: ukca_nmspec_mod.o |
||
| + | [FAIL] ! ukca_nmspec_mod.o : update task failed |
||
| + | |||
| + | You will notice that the file being pointed to is in the <code>cylc-run/</code> directory: |
||
| + | |||
| + | '''/home/ubuntu/cylc-run/u-ca024/share/fcm_make/preprocess-atmos/src/um/src/atmosphere/UKCA/ukca_nmspec_mod.F90''' |
||
| + | |||
| + | To view the exact line number referenced (line '''160''' in this case) you should open this file printing in the job.err file and then search for the same line in the equivalent file in your working copy. You may find that the line numbers are quite different - for instance line 160 in this file in <code>cylc-run/</code> directory is line 175 in the equivalent file in the working copy. |
||
==Solution to Task 4.2== |
==Solution to Task 4.2== |
||
Latest revision as of 09:22, 17 March 2022
UKCA Chemistry and Aerosol Tutorials at vn11.8
| Difficulty | HARD |
| Time to Complete | 2 or more hours |
| Video instructions | https://youtu.be/6XqYfW2qHKY?t=213 |
Remember to run mosrs-cache-password in your Rose suite and UM branch terminals.
What you will do in this tutorial
In this tutorial you will learn how to make the required changes in Rose and in UKCA to include new chemical tracers. As you learn how to do this you will add two new tracers to the UKCA tutorial suite you have already been running.
Task 4.1: Add new tracers into Rose
TASK4.1: Add in two new tracers in to slots 498 and 499 for the StratTrop chemistry scheme. The tracer in slot 498 will be called ALICE and the tracer in slot 499 will be called BOB. You should initialise these tracers to 1.000000e-15. You should also output these two tracers through the UPA stream in STASH as 3-hour means. Do not make any UKCA code changes at this stage.
Note: if you are working at UM versions prior to UMvn11.7, you will only be able to use tracer slots up to 256 (rather than 499 as used here). For more information see the vn10.9 Tutorials.
Make a branch
As UKCA is a framework, it must have the infrastructure around it to allow it to work, with the infrastructure being the UM itself. In this tutorial you will add in two new chemical tracers. This can be done in two steps, the Rose-side changes, and then the UKCA-side changes. Note that you cannot make the UKCA changes first.
For this tutorial you will need to make a new branch in the usual way. You should do this now.
You will have learned how to make UM branches in the NCAS-CMS FCM Tutorial. However, now you should use um.x_tr as the source of your branch.
For example, first you should make a ticket on the Met Office SRS Trac pages (login required) and then make the branch like so
fcm branch-create --type dev -k ticket_number your_branch_name fcm:um.x_tr@vn11.8
before checking-out your branch by
fcm checkout fcm:um.x_br/dev/[your MOSRS userid]/vn11.8_your_branch_name
More information on FCM can be found at the NCAS-CMS FCM pages or the FCM User Guide
You should
- ensure that you assign the ticket to yourself when you create it. The easiest way to do this is to click start work under the modify ticket option.
- When making changes always include your ticket number and MOSRS userid in your comment when you commit to your branch to enable the changes to be easily traced back to you. The easiest way to do this is to have each comment start with
#ticket_number userid. - Ensure that you select a Milestone of Not for builds. Usual changes should be given this as a milestone, or Hopefully. You should only change this to a particular UM release when you are working on it to commit to the trunk.
Metadata
In Rose, the GUI appearance is controlled by metadata, and it is possible to edit this metadata to add new variables to panels. We will not do this in this tutorial, but you will need to change where Rose looks for metadata, as the STASHmaster_A is also metadata.
Using your new metadata within Rose
Here you should click on the um app in Rose and change the meta setting from um-atmos/vn11.8 to
/path/to/vn11.8_your_branch_name/rose-meta/um-atmos/HEAD
Then click Apply and then save the suite.
Rose Changes
Include your branch for code changes
Although we haven't made any code changes yet, you will (at some point) need to include your branch in Rose so that the UKCA code changes can be picked-up and compiled. To do this go to fcm_make env Sources and add a new branch by clicking the plus symbol in the um_sources section. You should then put
/path/to/vn11.8_your_branch_name
(where /path/to/vn11.8_your_branch_name means the full linux path to your branch, e.g. /home/ukcatr01/vn11.8_UKCA_Tutorials etc.) if you wish to run from a working copy, and
branches/dev/[your MOSRS userid]/vn11.8_your_branch_name
if you want to run from the repository. You can specify specific revision numbers by putting @REV at the end of this line (where REV is the revision number, e.g. 12345 etc.).
Note that if you specify revision numbers you will need to change these each time you make a change and commit it. Another potential issue here is the use of a mirror for the UM code. On some systems the code is not taken directly from MOSRS, but from a local mirror of the full repository. This usually updates every 5 minutes or so, so to check whether it has updated or not, you should type the command
fcm info fcm:um.xm
and check the last committed revision. If it is the same or higher than your last revision then your code changes will be picked-up. If it is less than your revision your code changes will not be included in the compile. On the VM, this is not an issue as there is no mirror.
STASHmaster_A
In Rose-based jobs, adding new UKCA tracers is done through editing the STASHmaster_A file directly. Take a look at the STASHmaster_A file, found in
/path/to/vn11.8_your_branch_name/rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster_A
This is a large file, as it contains all the possible STASH items. Scroll down the file until you get to section 34, and you will see how the tracers are defined. The key entries to consider are highlighted in red. All entries are fixed-width, so when editing the file, ensure not to change the spacing, otherwise you will get errors.
#=============================================================================== # Section 34 UKCA Chemistry #=============================================================================== # 1| 1 | 34 | 1 |O3 MASS MIXING RATIO AFTER TIMESTEP | 2| 2 | 0 | 1 | 1 | 2 | 40 | 11 | 0 | 0 | 0 | 0 | 3| 000000000000000000100000111110 | 00000000000000000001 | 1 | 4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | 5| 0 | 2101 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | #
The meaning of these entries is:
First Line:
- 34: The STASH section, in this case Section 34, UKCA prognostics.
- 1: The STASH item, in this case, the first item in s34, ozone.
- O3 MASS MIXING RATIO AFTER TIMESTEP: The name of the field in the STASHmaster file. This can be anything descriptive, but it isn't used by the model, and is only considered in Rose (or e.g. Xconv).
Second Line:
- 2: The Space Code, which tells the UM whether the field should be held in the restart dump or not. For prognostics, this should be 2 (which means that they should be stored in the restart dump), but for diagnostics that are only passed to output files, this should be 0.
Third Line:
- 000000000000000000100000111110: These are Options Codes which determine which scheme the STASH field (i.e. tracer in this case) are valid for. These are counted from the right, n1 to n30. Only some of these are used, and have the following meaning:
- n1: Age-of-air tracer (ageair) or aerosol tracers
- n2: Standard Tropospheric Chemistry scheme using the Backward Euler solver (trop)
- n3: Regional Air Quality Chemistry scheme using the Backward Euler solver (raq)
- n4: Tropospheric Chemistry with Isoprene scheme using the Newton-Raphson solver (tropisop)
- n5: Stratospheric-Tropospheric Chemistry using the Newton-Raphson solver (strattrop, also know as CheST)
- n6: Standard Stratospheric Chemistry using the Newton-Raphson solver (strat)
- n7: Offline-oxidants scheme (to drive GLOMAP-mode) using the Newton-Raphson solver (offline)
- n8: Offline-oxidants scheme (to drive GLOMAP-mode) using the Newton-Raphson solver (offline_be)
- n12: CRI-Strat scheme using the Newton-Raphson solver (cristrat)
Therefore, looking at the option code for ozone (s34i001), it is valid for the trop, raq, tropisop, strattrop, strat, and cristrat chemistry schemes.
Fifth Line:
- 2101: This is the PP Field Code. Existing UKCA fields will have different numbers for these, but for all new fields, you should set this number to 0.
So to make a STASH entry for a single tracer called ALICE in slot s34i498 that is only valid in the StratTrop scheme, the STASHmaster_A entry would look like
# 1| 1 | 34 | 498 |ALICE MASS MIXING RATIO AFTER TSTEP | 2| 2 | 0 | 1 | 1 | 2 | 40 | 11 | 0 | 0 | 0 | 0 | 3| 000000000000000000000000010000 | 00000000000000000001 | 1 | 4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | 5| 0 | 0 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | #
Remember: You need to add 2 tracers, ALICE in s34i498 and BOB in s34i499.
Section 51
All STASH entries in Section 34 need a corresponding pressure-level field to be added to section 51. The equivalent STASHmaster_A entry for s51 ozone is:
# #=============================================================================== # Section 51 UKCA Chemistry on pressure levels #=============================================================================== # 1| 1 | 51 | 1 |O3 MASS MIXING RATIO ON PRESS LEVS | 2| 0 | 0 | 1 | 1 | 3 | 1 | 2 | 0 | 0 | 0 | 1 | 3| 000000000000000000000000111110 | 00000000000000000001 | 3 | 4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 | 5| 0 | 2101 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | #
You will notice that there are a number of changes to various codes, and this is to do with defining the grid that the diagnostic is valid on, whether it can be held in the dump, etc. For more information on what these codes mean, please see Appendix C in UMDP C4, which can be obtained from the SRS here (password required). When making a new entry, the easiest thing to do is copy an existing entry and make the required changes to the item, name, PP field code, and option codes.
Remember: You need to add 2 tracers, ALICE in s34i498 and BOB in s34i499 (and section 51 for pressure level equivalents).
Help text
You should also add appropriate help text into the vn11.8_your_branch_name/rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster-meta.conf file, e.g.
[stashmaster:code(34498)] description=ALICE MASS MIXING RATIO AFTER TIMESTEP help=UKCA Tutiorial Tracer ALICE Mass Mixing Ratio in kg/kg(Air)
and
[stashmaster:code(51498)]
description=ALICE MASS MIX RATIO ON PRESS LEVS
help=UKCA Tutiorial Tracer ALICE Mass Mixing Ratio in kg/kg(Air)
=on pressure levels
Remember: You need to add 2 tracers, ALICE in s34i498 and BOB in s34i499 (and section 51 for pressure level equivalents).
You should make these changes to STASHmaster_A and STASHmaster-meta.conf, and then fcm commit these changes. This is important, otherwise the running suite will not be able to see the changes you have made once you have included your new STASHmaster_A file in Rose.
Use your new STASHmaster file in Rose
Before you make these changes you should first save and close your suite.
To get Rose to recognise your STASHmaster file, you will need to make a number of changes:
- Make sure to point the um metadata to your branch (if it isn't already) by editing the um meta path (you should already be doing this after following the instructions in the metadata section above).
- You need to open the
$HOME/roses/[SUITE-ID]/app/um/rose-app.conffile in a text editor (e.g. vim, emacs, nedit etc.) and add the lineSTASHMASTER=STASHmasterinside the[env]block near the top of the file. - You need to open the
$HOME/roses/[SUITE-ID]/rose-suite.conffile in a text editor and add the following lines at the top of the file:
[file:app/um/file/STASHmaster] source=fcm:um.xm_br/dev/[your MOSRS userid]/vn11.8_your_branch_name/rose-meta/um-atmos/HEAD/etc/stash/STASHmaster@HEAD
- You need to ensure that you have committed your STASHmaster_A changes, otherwise your new tracer(s) will not be seen at run-time during the reconfiguration or atmosphere steps.
- For the VM only it is also possible to specify the full path to the STASHmaster directory. This is because the suite runs on the same machine that the branch is on.
[file:app/um/file/STASHmaster] source=/home/[your VM userid]/src/vn11.8_your_branch_name/rose-meta/um-atmos/HEAD/etc/stash/STASHmaster
You may wish to now fcm commit your suite changes. You can now re-open your suite.
When running your suite, if you get an error similar to:
[INFO] create: app/um/file [FAIL] file:app/um/file/STASHmaster=source=fcm:um.xm_br/dev/[your MOSRS userid]/vn11.8_your_branch_name/rose-meta/um-atmos/HEAD/etc/stash/STASHmaster@HEAD: bad or missing value
you should try running mosrs-cache-password again to cache your MOSRS credentials.
Initialise your new tracer(s)
Your new tracer(s) is a prognostic variable, as therefore the UM requires prior knowledge of the values it has to be able to run properly. To do this you need to go to um namelist Reconfiguration and Ancillary Control Configure ancils and initialise dump fields. Here you will see a list of fields. You can change the order of these by e.g. STASH number, as this often makes it easier to view.
To add new fields, right click anywhere within the table and click Add new section. This will make a new entry with index 1, if you add another entry, it will be given index 2 etc.. Right-click on this and click on View namelist:items(X) (where X is the index number). This will open a new tab where you can select the STASH section and item for the field you want to initialise using the drop-down menus. Some UKCA tracers need to be given full 3D initial conditions in ancillary file format. However, for this example we will initialise the tracer to a small number, . To do this, select the radio button for Set to a specified constant value and put 1.000000e-15 in the dialog box labelled user_prog_rconst at the bottom of the panel.
You will need to do this for each of the tracers you are adding, and you can add multiple prognostics per entry. You can also clone existing entries to speed up the process. When you have finished, you will need to go to the STASH Requests panel (see the STASH tutorial) and run the TidyStashTransform macro to correctly generate the required index value for each tracer initialisation.
Output your new tracers
To output your tracers, go to the STASH panel and output your tracers as described in the What is STASH? tutorial.
Remember: You need to add 2 tracers, ALICE in s34i498 and BOB in s34i499.
Run your suite
Now that you have made the required changes to Rose and in your branch, please run your suite. You may find that it fails on the fcm_make, recon, or atmos jobs. To find the errors and see output, you should either right-click the failed task in Gcylc and view the output, or go to the
/home/[your VM userid]/cylc-run/[SUITE-ID]
directory. Within this there are a number of directories. To see the log files, cd into
log/job/1/[JOB NAME]/NN
and view the job.err or job.out files. When the suite successfully runs, the fieldsfile output can be found in
/home/[your VM userid]/cylc-run/[SUITE-ID]/work/1/atmos/
Viewing output in Xconv
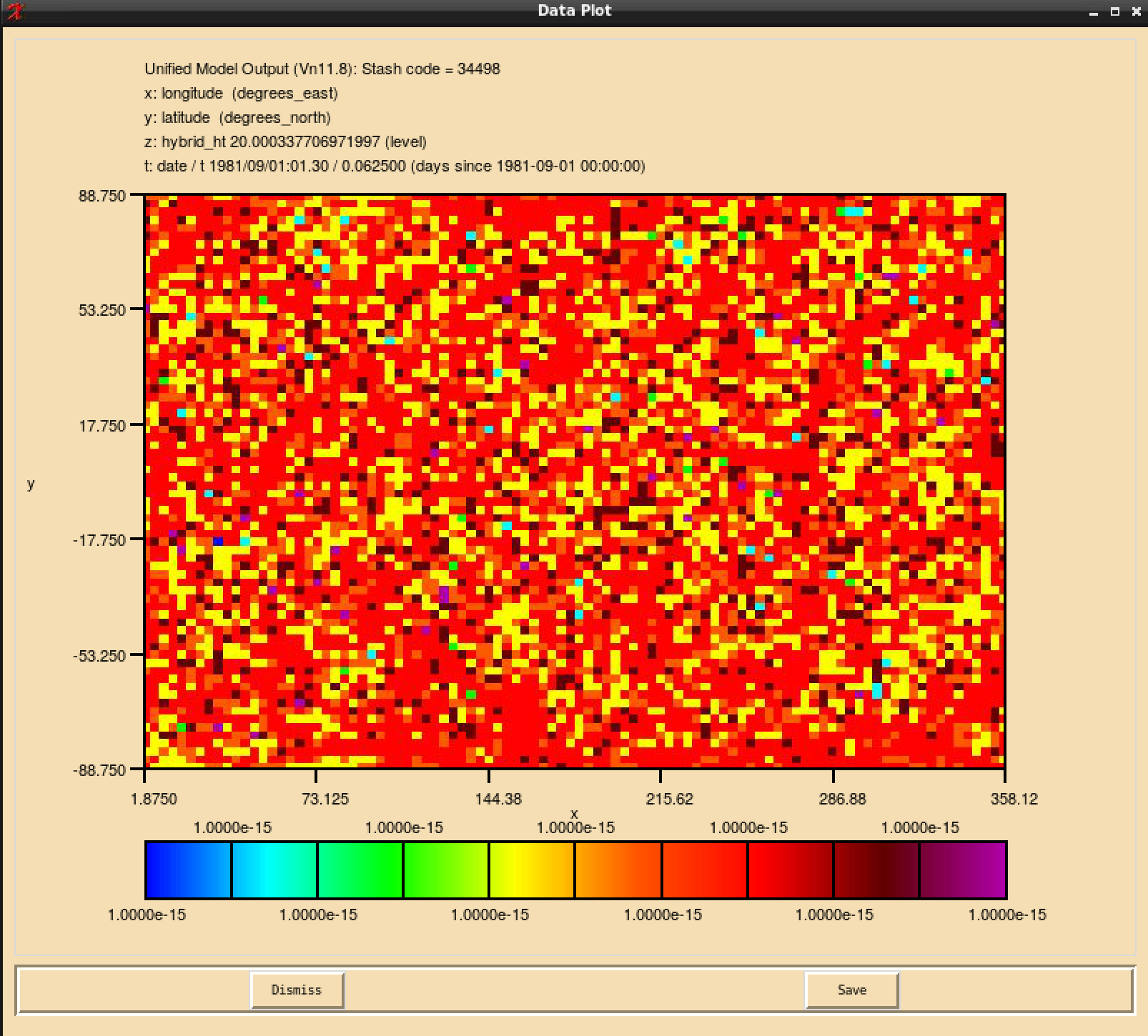
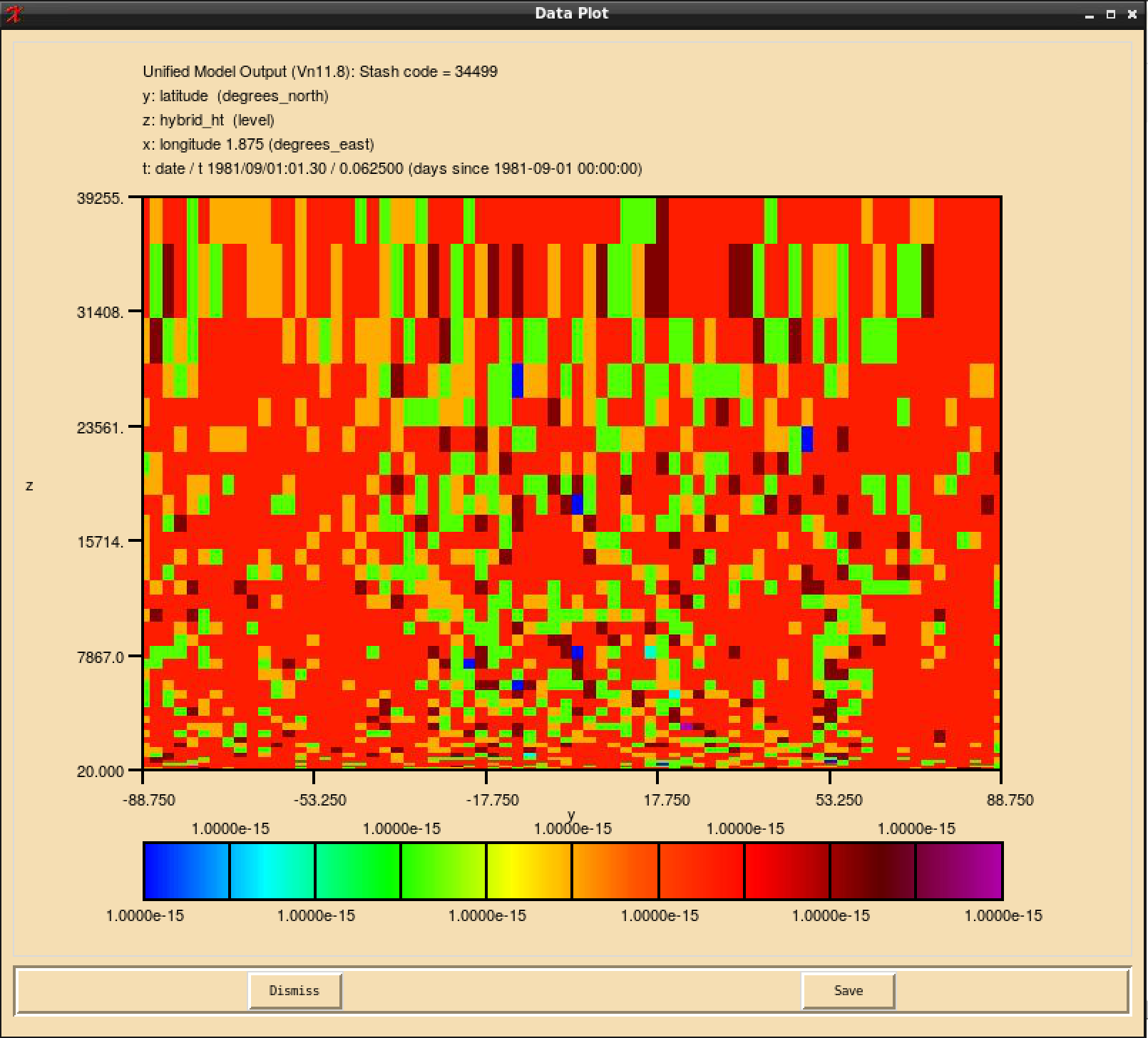
If you open the .pa file in Xconv, you should now see the following if you have outputted both the section 34 and section 51 fields:
4 : 96 72 27 1 Stash code = 51498 5 : 96 72 27 1 Stash code = 51499 11 : 96 72 38 1 Stash code = 34498 12 : 96 72 38 1 Stash code = 34499
Note here that these fields are not named ALICE and BOB, or however you have described them in your STASHmaster_A file. They are instead labelled by the unique STASH code identifier that you have defined for them. This is working as expected.
- Optional: It is possible (although not necessary) to tell Xconv to read the STASHmaster_A file from your branch. To do this, in Xconv go to Setup Select STASH Master Files and then navigate to the location of the file in your branch's working copy in the left-hand side of the panel. When you have located it, highlight it and click Add. Then click Apply, Save, and Dismiss. Now Xconv will list the names of your species as you have called them in your STASHmaster_A file.
Numerical Noise
When your suite has run successfully, and you view the fields for your new tracer(s) in Xconv, you may find that they have a speckled appearance, rather than being constant. This is due to numerical noise that has been introduced as the tracers go through the UM timesteps. Once the field is converted to a 32-bit pp-file this should disappear. It will also cease to become apparent when emissions and reactions are applied to these tracers.
Solution to Task4.1
You were given the task:
- Add in two new tracers in to slots 498 and 499 for the StratTrop chemistry scheme. The tracer in slot 498 will be called ALICE and the tracer in slot 499 will be called BOB. You should initialise these tracers to
1.000000e-15. You should also output these two tracers through the UPA stream in STASH as 3-hour means. Do not make any UKCA code changes at this stage.
For a working Rose suite that has completed this task, please see
- vm:
u-ca024@181051
The specific Rose changes made are:
vm:
Index: app/um/rose-app.conf =================================================================== --- app/um/rose-app.conf (revision 180902) +++ app/um/rose-app.conf (revision 181051) @@ -1,4 +1,4 @@ -meta=um-atmos/vn11.8 +meta=/home/vagrant/src/vn11.8_UKCA_Tutorial_Solns/rose-meta/um-atmos/HEAD [command] default=um-atmos @@ -27,6 +27,7 @@ RECON_STDOUT_FILE=pe_output/atmos.fort6.pe RUNID=atmos SPECTRAL_FILE_DIR=$UMDIR/vn$VN/ctldata/spectral/ga7 +STASHMASTER=STASHmaster UM_THREAD_LEVEL=MULTIPLE [file:$DATAM] @@ -893,6 +894,19 @@ !!user_prog_ancil_stash_req= !!user_prog_rconst=0.0 +[namelist:items(e897a2fb)] +ancilfilename= +domain=1 +!!interval=0 +l_ignore_ancil_grid_check= +!!netcdf_varname= +!!period=1 +source=6 +stash_req=34498,34499 +update_anc=.false. +!!user_prog_ancil_stash_req=0 +user_prog_rconst=1.000000e-15 + [namelist:items(e8e484ca)] ancilfilename= domain=1 @@ -3658,6 +3672,22 @@ tim_name='T3HMN' use_name='UPA' +[namelist:umstash_streq(34498_e7e57cc6)] +dom_name='DALLTH' +isec=34 +item=498 +package= +tim_name='T3HMN' +use_name='UPA' + +[namelist:umstash_streq(34499_07c8a074)] +dom_name='DALLTH' +isec=34 +item=499 +package= +tim_name='T3HMN' +use_name='UPA' + [namelist:umstash_streq(34968_49e5e12c)] dom_name='DALLTH' isec=34 @@ -3770,6 +3800,22 @@ tim_name='T3HMN' use_name='UPA' +[namelist:umstash_streq(51498_ccb8d5a7)] +dom_name='DP27CCM' +isec=51 +item=498 +package= +tim_name='T3HMN' +use_name='UPA' + +[namelist:umstash_streq(51499_5c041195)] +dom_name='DP27CCM' +isec=51 +item=499 +package= +tim_name='T3HMN' +use_name='UPA' + [namelist:umstash_streq(51999_8b73e62f)] dom_name='DP27CCM' isec=51 Index: app/fcm_make/rose-app.conf =================================================================== --- app/fcm_make/rose-app.conf (revision 180902) +++ app/fcm_make/rose-app.conf (revision 181051) @@ -43,4 +43,4 @@ thread_utils=false timer_version=3A um_rev=$BASE_UM_REV -um_sources= +um_sources=branches/dev/lukeabraham/vn11.8_UKCA_Tutorial_Solns@93557 Index: rose-suite.conf =================================================================== --- rose-suite.conf (revision 180902) +++ rose-suite.conf (revision 181051) @@ -1,3 +1,6 @@ +[file:app/um/file/STASHmaster] +source=fcm:um.xm_br/dev/lukeabraham/vn11.8_UKCA_Tutorial_Solns/rose-meta/um-atmos/HEAD/etc/stash/STASHmaster@HEAD + [jinja2:suite.rc] BUILD=true OFFLINE=false
These ARCHER differences can be found in the file Tutorials/vn11.8/worked_solutions/Task04.1/Task04.1_rose.patch.
The specific UM changes made are:
Index: rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster-meta.conf
===================================================================
--- rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster-meta.conf (revision 93540)
+++ rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster-meta.conf (revision 93557)
@@ -19390,6 +19390,14 @@
help=Coarse Mode (Soluble) Sodium Nitrate Aerosol Mass Mixing
=Ratio in kg/kg(Air)
+[stashmaster:code(34498)]
+description=ALICE MASS MIXING RATIO AFTER TIMESTEP
+help=UKCA Tutorial Tracer ALICE Mass Mixing Ratio in kg/kg(Air)
+
+[stashmaster:code(34499)]
+description=BOB MASS MIXING RATIO AFTER TIMESTEP
+help=UKCA Tutorial Tracer BOB Mass Mixing Ratio in kg/kg(Air)
+
[stashmaster:code(34866)]
description=NH4 PARTIAL VOLUME CONC (AIT-SOL)
help=Component partial volume of ammonium for the Aitken
@@ -27596,6 +27604,16 @@
=Mass Mixing Ratio
=in kg/kg(Air) on pressure levels
+[stashmaster:code(51498)]
+description=ALICE MASS MIXING RATIO AFTER TIMESTEP
+help=UKCA Tutorial Tracer ALICE Mass Mixing Ratio in kg/kg(Air)
+ =on pressure levels
+
+[stashmaster:code(51499)]
+description=BOB MASS MIXING RATIO AFTER TIMESTEP
+help=UKCA Tutorial Tracer BOB Mass Mixing Ratio in kg/kg(Air)
+ =on pressure levels
+
[stashmaster:code(51872)]
description=RTX22O2 MASS MIX RATIO ON PRESS LEVS
help=Peroxy radicals from BPINENE oxidation with 17 C-H
Index: rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster_A
===================================================================
--- rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster_A (revision 93540)
+++ rose-meta/um-atmos/HEAD/etc/stash/STASHmaster/STASHmaster_A (revision 93557)
@@ -18682,6 +18682,18 @@
4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
5| 0 | 2351 | 0 | 65 | 0 | 0 | 0 | 0 | 0 |
#
+1| 1 | 34 | 498 |ALICE MASS MIXING RATIO AFTER TSTEP |
+2| 2 | 0 | 1 | 1 | 2 | 40 | 11 | 0 | 0 | 0 | 0 |
+3| 000000000000000000000000010000 | 00000000000000000001 | 1 |
+4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
+5| 0 | 0 | 0 | 65 | 0 | 0 | 0 | 0 | 0 |
+#
+1| 1 | 34 | 499 |BOB MASS MIXING RATIO AFTER TSTEP |
+2| 2 | 0 | 1 | 1 | 2 | 40 | 11 | 0 | 0 | 0 | 0 |
+3| 000000000000000000000000010000 | 00000000000000000001 | 1 |
+4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
+5| 0 | 0 | 0 | 65 | 0 | 0 | 0 | 0 | 0 |
+#
1| 1 | 34 | 866 |NH4 PARTIAL VOLUME CONC (AIT-SOL) |
2| 2 | 0 | 1 | 1 | 2 | 40 | 11 | 0 | 0 | 0 | 0 |
3| 100000000000000000005000000000 | 00000000000000000001 | 3 |
@@ -29005,6 +29017,18 @@
4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
5| 0 | 2252 | 0 | 8 | 0 | 0 | 0 | 0 | 0 |
#
+1| 1 | 51 | 498 |ALICE MMR ON PRESSURE LEVELS |
+2| 0 | 0 | 1 | 1 | 3 | 1 | 2 | 0 | 0 | 0 | 1 |
+3| 000000000000000000000000010000 | 00000000000000000001 | 3 |
+4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
+5| 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 |
+#
+1| 1 | 51 | 499 |BOB MMR ON PRESSURE LEVELS |
+2| 0 | 0 | 1 | 1 | 3 | 1 | 2 | 0 | 0 | 0 | 1 |
+3| 000000000000000000000000010000 | 00000000000000000001 | 3 |
+4| 1 | 0 | -99 -99 -99 -99 -99 -99 -99 -99 -99 -99 |
+5| 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 |
+#
1| 1 | 51 | 872 |RTX22O2 MASS MIX RATIO ON PRESS LEVS|
2| 0 | 0 | 1 | 1 | 3 | 1 | 2 | 0 | 0 | 0 | 1 |
3| 000000000000000000800000000000 | 00000000000000000001 | 3 |
These differences can be found in the file Tutorials/vn11.8/worked_solutions/Task04.1/Task04.1_code.patch.
If you open the .pa file in Xconv, you should see the following fields:
4 : 96 72 27 1 Stash code = 51498 5 : 96 72 27 1 Stash code = 51499 11 : 96 72 38 1 Stash code = 34498 12 : 96 72 38 1 Stash code = 34499
Sample output from this task can be found at Tutorials/vn11.8/sample_output/Task04.1/atmosa.pa19810901_00.
Task 4.2: Add new tracers into UKCA
TASK4.2: In UKCA, make the code changes required to include these species, ALICE and BOB, into the StratTrop chemical mechanism. You should set the conversion factor for each of these to 1.0. In the nm_spec array you should put these into slots 498 for ALICE and 499 for BOB.
Required UKCA Code Changes
Note that at some older UKCA versions it was necessary to make the UKCA changes in parallel with the UM-side changes covered above.
ukca_nmspec_mod.F90
Inside UKCA there is a master mapping array that tells the UKCA code which transported species are placed into a particular item number in section 34. At vn11.8 these are contained in the first 499 slots.
This array is called nm_spec, and it can be found in ukca_nmspec_mod.F90. At vn11.8, it looks like this:
nm_spec(1:a_max_ukcavars) = (/ & 'O3 ','NO ','NO3 ','NO2 ','N2O5 ', & 'HO2NO2 ','HONO2 ','H2O2 ','CH4 ','CO ', & !10 'HCHO ','MeOOH ','HONO ','C2H6 ','EtOOH ', & 'MeCHO ','PAN ','C3H8 ','n-PrOOH ','i-PrOOH ', & !20 'EtCHO ','Me2CO ','MeCOCH2OOH','PPAN ','MeONO2 ', & 'O3_S ','C5H8 ','ISOOH ','ISON ','MACR ', & !30 'MACROOH ','MPAN ','HACET ','MGLY ','NALD ', & 'HCOOH ','MeCO3H ','MeCO2H ','H2O ','ISO2 ', & !40 'Cl ','ClO ','Cl2O2 ','OClO ','Br ', & 'BrO ','BrCl ','BrONO2 ','N2O ','HCl ', & !50 'HOCl ','HBr ','HOBr ','ClONO2 ','CFCl3 ', & 'CF2Cl2 ','MeBr ','N ','O(3P) ','MACRO2 ', & !60 'C2H4 ','C4H10 ','oXYLENE ','TBUT2ENE ','APINENE ', & 'BPINENE ','C2H2 ','BENZENE ','CH2Br2 ','H2 ', & !70 'DMS ','SO2 ','H2SO4 ','MSA ','DMSO ', & 'NH3 ','CS2 ','COS ','H2S ','H ', & !80 'OH ','HO2 ','MeOO ','EtOO ','MeCO3 ', & 'n-PrOO ','i-PrOO ','EtCO3 ','MeCOCH2OO ','MeOH ', & !90 'Monoterp ','Sec_Org ','C3H6 ','SO3 ','C4H9OOH ', & 'MEK ','TOLUENE ','NO2 ','BrO ','HCl ', & !100 'Nuc_SOL_ND','Nuc_SOL_SU','Ait_SOL_ND','Ait_SOL_SU','Ait_SOL_BC', & 'Ait_SOL_OC','Acc_SOL_ND','Acc_SOL_SU','Acc_SOL_BC','Acc_SOL_OC', & !110 'Acc_SOL_SS','Acc_SOL_DU','Cor_SOL_ND','Cor_SOL_SU','Cor_SOL_BC', & 'Cor_SOL_OC','Cor_SOL_SS','Cor_SOL_DU','Ait_INS_ND','Ait_INS_BC', & !120 'Ait_INS_OC','Acc_INS_ND','Acc_INS_DU','Cor_INS_ND','Cor_INS_DU', & 'Nuc_SOL_OC','Ait_SOL_SS','Nuc_SOL_SO','Ait_SOL_SO','Acc_SOL_SO', & !130 'Cor_SOL_SO','Nuc_SOL_NH','Ait_SOL_NH','Acc_SOL_NH','Cor_SOL_NH', & 'Nuc_SOL_NT','Ait_SOL_NT','Acc_SOL_NT','Cor_SOL_NT','EtOH ', & !140 'i-PrOH ','n-PrOH ','HOCH2CHO ','HOC2H4OOH ','EtCO3H ', & !145 'HOCH2CO3H ','NOA ','EtONO2 ','PASSIVE O3','AGE OF AIR', & !150 'i-PrONO2 ','MeO2NO2 ','HOC2H4NO3 ','PHAN ','MeSCH2OO ', & !155 'MeS ','MeSO ','MeSO2 ','MeSO3 ','MSIA ', & !160 'CARB14 ','CARB17 ','CARB11A ','CARB7 ','CARB10 ', & !165 'CARB13 ','CARB16 ','UCARB10 ','CARB3 ','CARB6 ', & !170 'CARB9 ','CARB12 ','CARB15 ','UCARB12 ','NUCARB12 ', & !175 'UDCARB8 ','UDCARB11 ','UDCARB14 ','TNCARB26 ','TNCARB10 ', & !180 'RN10NO3 ','RN13NO3 ','RN16NO3 ','RN19NO3 ','RA13NO3 ', & !185 'RA16NO3 ','RA19NO3 ','RTX24NO3 ','RN10OOH ','RN13OOH ', & !190 'RN16OOH ','RN19OOH ','RN8OOH ','RN11OOH ','RN14OOH ', & !195 'RN17OOH ','RU14OOH ','RU12OOH ','RU10OOH ','NRU14OOH ', & !200 'NRU12OOH ','RN9OOH ','RN12OOH ','RN15OOH ','RN18OOH ', & !205 'NRN6OOH ','NRN9OOH ','NRN12OOH ','RA13OOH ','RA16OOH ', & !210 'RA19OOH ','RTN28OOH ','NRTN28OOH ','RTN26OOH ','RTN25OOH ', & !215 'RTN24OOH ','RTN23OOH ','RTN14OOH ','RTN10OOH ','RTX28OOH ', & !220 'RTX24OOH ','RTX22OOH ','NRTX28OOH ','RAROH14 ','RAROH17 ', & !225 'RU12PAN ','RTN26PAN ','TNCARB12 ','TNCARB11 ','RTN23NO3 ', & !230 'CCARB12 ','TNCARB15 ','RCOOH25 ','TXCARB24 ','TXCARB22 ', & !235 'RN9NO3 ','RN12NO3 ','RN15NO3 ','RN18NO3 ','RU14NO3 ', & !240 'RTN28NO3 ','RTN25NO3 ','RTX28NO3 ','RTX22NO3 ','AROH14 ', & !245 'ARNOH14 ','AROH17 ','ARNOH17 ','ANHY ','Acc_SOL_NN', & !250 'Cor_SOL_NN','XXX ','XXX ','XXX ','XXX ', & !255 'XXX ','XXX ','XXX ','XXX ','XXX ', & !260 'XXX ','XXX ','XXX ','XXX ','XXX ', & !265 'XXX ','XXX ','XXX ','XXX ','XXX ', & !270 'XXX ','XXX ','XXX ','XXX ','XXX ', & !275 'XXX ','XXX ','XXX ','XXX ','XXX ', & !280 'XXX ','XXX ','XXX ','XXX ','XXX ', & !285 'XXX ','XXX ','XXX ','XXX ','XXX ', & !290 'XXX ','XXX ','XXX ','XXX ','XXX ', & !295 'XXX ','XXX ','XXX ','XXX ','XXX ', & !300 'XXX ','XXX ','XXX ','XXX ','XXX ', & !305 'XXX ','XXX ','XXX ','XXX ','XXX ', & !310 'XXX ','XXX ','XXX ','XXX ','XXX ', & !315 'XXX ','XXX ','XXX ','XXX ','XXX ', & !320 'XXX ','XXX ','XXX ','XXX ','XXX ', & !325 'XXX ','XXX ','XXX ','XXX ','XXX ', & !330 'XXX ','XXX ','XXX ','XXX ','XXX ', & !335 'XXX ','XXX ','XXX ','XXX ','XXX ', & !340 'XXX ','XXX ','XXX ','XXX ','XXX ', & !345 'XXX ','XXX ','XXX ','XXX ','XXX ', & !350 'XXX ','XXX ','XXX ','XXX ','XXX ', & !355 'XXX ','XXX ','XXX ','XXX ','XXX ', & !360 'XXX ','XXX ','XXX ','XXX ','XXX ', & !365 'XXX ','XXX ','XXX ','XXX ','XXX ', & !370 'XXX ','XXX ','XXX ','XXX ','XXX ', & !375 'XXX ','XXX ','XXX ','XXX ','XXX ', & !380 'XXX ','XXX ','XXX ','XXX ','XXX ', & !385 'XXX ','XXX ','XXX ','XXX ','XXX ', & !390 'XXX ','XXX ','XXX ','XXX ','XXX ', & !395 'XXX ','XXX ','XXX ','XXX ','XXX ', & !400 'XXX ','XXX ','XXX ','XXX ','XXX ', & !405 'XXX ','XXX ','XXX ','XXX ','XXX ', & !410 'XXX ','XXX ','XXX ','XXX ','XXX ', & !415 'XXX ','XXX ','XXX ','XXX ','XXX ', & !420 'XXX ','XXX ','XXX ','XXX ','XXX ', & !425 'XXX ','XXX ','XXX ','XXX ','XXX ', & !430 'XXX ','XXX ','XXX ','XXX ','XXX ', & !435 'XXX ','XXX ','XXX ','XXX ','XXX ', & !440 'XXX ','XXX ','XXX ','XXX ','XXX ', & !445 'XXX ','XXX ','XXX ','XXX ','XXX ', & !450 'XXX ','XXX ','XXX ','XXX ','XXX ', & !455 'XXX ','XXX ','XXX ','XXX ','XXX ', & !460 'XXX ','XXX ','XXX ','XXX ','XXX ', & !465 'XXX ','XXX ','XXX ','XXX ','XXX ', & !470 'XXX ','XXX ','XXX ','XXX ','XXX ', & !475 'XXX ','XXX ','XXX ','XXX ','XXX ', & !480 'XXX ','XXX ','XXX ','XXX ','XXX ', & !485 'XXX ','XXX ','XXX ','XXX ','XXX ', & !490 'XXX ','XXX ','XXX ','XXX ','XXX ', & !495 'XXX ','XXX ','XXX ','XXX ' & !499 /)
This array is case sensitive and each string is a fixed length of 10 characters, with the whitespace being made up of spaces and not another type of white space (e.g. tabs).
When adding new tracers you should not overwrite tracers that are already in use by the chemistry scheme that you are using (e.g. StratTrop etc.), and it is best to also take care and avoid tracers in use by other schemes, if possible. You can see the tracers used by each scheme by looking in the chch_defs array at the top of the ukca_chem_master.F90 module.
Here you should put a new species, 'ALICE ', into tracer slot 498 (currently specified as 'XXX ', which means that it is empty and is not in use by any scheme at the moment).
Remember: You need to add 2 tracers, ALICE in s34i498 and BOB in s34i499.
ukca_chem_master.F90
We will go through changes that need to be made for the StratTrop chemistry scheme (defined in the file ukca_chem_master.F90, using the code st), but equivalent changes would need to be made for others, e.g. TropIsop or Strat. For this example we will only refer to this scheme.
In this file the list of species used in chemistry is defined by the array chch_defs_master. The list of species is held in a derived type, of the format:
chch_t1( N,'SPECIES ', X,'TYPE ','FAMILY ', D, W, SCHEME, QUALIFIER, DISQUALIFIER, VN), &
Where:
Nis an integer. The value should be incremented for each new species that is added.'SPECIES 'is the name of the species, e.g. 'O3 'Xnumber of odd atoms'TYPE 'describes whether the species is- a tracer ('TR ')
- a steady-state species ('SS ')
- a constant ('CT ')
- a constant field ('CF ').
- For SS, CT, and CF, special code will need to be added.
'FAMILY 'is the family that the species belongs to. This field is not currently used.Dis 1 if the species is dry-deposited, and 0 otherwise.Wis 1 if the species is wet-deposited, and 0 otherwise.SCHEMEdefines the chemistry scheme used, e.g.:st= Stratosphere-Troposphere schemet= Troposphere schemes= Stratosphere schemer= RAQ schemeol= Offline-oxidants schemeti= Troposphere-Isoprene schemecs= CRI-Strat scheme
- If the same species is used in multiple schemes these are added, e.g.
st+s+ti+csetc.
QUALIFIER(andDISQUALIFIER) defines lines to add (or remove) on top of the base chemistry scheme depending on if certain additional options have been chosen, e.g.- 0 = no qualifier/disqualifer used
a= Aerosol chemistry additions required to run GLOMAP-modeth= Tropospheric heterogeneous reactionshp= Heterogeneous PSC chemistryes= Extended stratospheric reactionsrn= RO2 a non-transported prognostic (StratTrop chemistry only)rp= RO2 permutation chemistry (StratTrop chemistry only)
- As with the
SCHEMEcodes, these are also additive if required, e.g.a+thetc.
VNgives the model version the addition is added at. This is future functionality that will allow for multiple rates to be included in the source-code. The currently used version is111, so please continue to use this for this Tutorial.
Therefore to add-in the ALICE species, we should insert a line similar to this one
chch_t1(272,'ALICE ',1,'TR ',' ',0,0,st,0,0,111), &
into chch_defs_master, remembering to also increment the size of this array, given by n_chch_master
Remember: You need to add 2 tracers, ALICE in s34i498 and BOB in s34i499.
ukca_config_defs_mod.F90
In ukca_config_defs_mod.F90 you will find code blocks which set values for various parameters depending on the scheme used. In this example you will need to scroll down to the section which defines the StratTrop (also known as CheST) chemistry, which is located in the Stratospheric Chemistry section and is referenced by using the ukca_config%l_ukca_strattrop logical.
The IF block tests against ukca_config%l_ukca_strattrop and ukca_config%l_ukca_achem (which determines whether or not you require the additional chemistry used to drive the GLOMAP-mode aerosol scheme). You should increment the value of n_chem_tracers by the number of chemical tracers that you are adding in both sections of the IF block, as this tracer is not defined to be for aerosol chemistry only. If your additional tracers are aerosol chemistry additions then you should increment the value of n_aero_tracers.
If you are adding to a different chemistry scheme then you will need to make these changes accordingly.
Remember: You need to add 2 tracers, ALICE in s34i498 and BOB in s34i498.
ukca_constants.F90
The unit of the tracers is kg(species)/kg(air) (i.e. mass mixing ratio, or mmr), but inside UKCA these species are converted to volume mixing ratio (or vmr). To enable UKCA to do this you will need to add the conversion factor(s) for your new tracer(s) into the code. This is done in the ukca_constants.F90 module.
Open this file and add the required conversion factor(s). The naming convention for these is M_species is the molecular mass of the new species in g/mol, and C_species for the converson factor from vmr to mmr (calculated as M_species/M_air, where M_air=28.97). In actuality, only the C_species listing is always required, although you may need to add the M_species value later if you are emitting into the new species that you are adding.
For example:
REAL, PARAMETER :: C_ALICE = 1.0000
Remember: You need to add 2 tracers, ALICE in s34i498 and BOB in s34i499.
ukca_cspecies.F90
After you have added in the C_species conversion factors, you will need to tell UKCA to use them for your species. To do this you need to edit the ukca_cspecies.F90 module, which contains code which constructs the c_species array of conversion factors for the advected tracers. This contains a subroutine called ukca_calc_cspecies which has a long block of code that you need to edit to add an entry like this:
WHERE (advt == 'ALICE ') c_species = C_ALICE
The advt array is automatically generated by UKCA at run-time from the chch_defs_master chemistry scheme definition you edited earlier, so your new tracer(s) will exist within it. You need to add in a new line for each of your tracers which sets the value of the c_species array to your individual C_species parameter. Add the line(s) in at the end of the block.
As the C_species conversion factors are defined in the ukca_constants module you will need to explicitly add them to the
USE ukca_constants, ONLY: ...
statement at the top of the module.
Remember: You need to add 2 tracers, ALICE in s34i498 and BOB in s34i499.
Run your suite again
Now that you have made the required changes to Rose and in your branch, please add your branch to your suite in Rose and run your suite again. To include the branch in your suite add it to the fcm_make env Sources panel. In the um_sources entry click the green cross (+) and put your branch in the box like
branches/dev/[your MOSRS userid]/vn11.8_your_branch_name
This will take the latest revision from the repository that you have checked-in (but not any code that you have not committed). You can also specify a fixed revision for a branch, e.g.
branches/dev/[your MOSRS userid]/vn11.8_your_branch_name@123456
In this case you will need to remember to update your branch revision number every time you commit a change.
You could also point to a working copy which will take the code as it is on the disk, including any code not committed to the repository, e.g.
/home/[your VM userid]/vn11.8_your_branch_name
and here you should not specify any revision numbers.
Failures and Debugging
You may find that it fails on the fcm_make, recon, or atmos jobs. To find the errors and see output, you should either right-click the failed task in Gcylc and view the output, or go to the
/home/[your VM userid]/cylc-run/[SUITE-ID]
directory. Within this there are a number of directories. To see the log files, cd into
log/job/1/[JOB NAME]/NN
and view the job.err or job.out files. When the suite successfully runs, the fieldsfile output can be found in
/home/[your VM userid]/cylc-run/[SUITE-ID]/work/1/atmos/
If you have errors in the fcm_make task it will give a line number in the Fortran file for where the error is. However, this line number will not be for your branch in your working copy, e.g.
/home/[your VM userid]/vn11.8_your_branch_name
but will instead be for code inside your cylc-run directory.
Example
If we consider the following error:
[FAIL] mpif90 -oo/ukca_nmspec_mod.o -c -I./include -I/home/vagrant/cylc-run/vn11.8_prebuilds/share/fcm_make_vm_gnu_um_safe_omp/build-atmos/include -fdefault-integer-8 -fdefault-real-8 -fdefault-double-8 -std=f2008ts -Wtabs -I/home/vagrant/umdir/gcom/default/vm_gfortran_mpich3/build/include -O2 -Werror -fopenmp -I/usr/include -I/usr/include /home/ubuntu/cylc-run/u-ca024/share/fcm_make/preprocess-atmos/src/um/src/atmosphere/UKCA/ukca_nmspec_mod.F90 # rc=1 [FAIL] /home/ubuntu/cylc-run/u-ca024/share/fcm_make/preprocess-atmos/src/um/src/atmosphere/UKCA/ukca_nmspec_mod.F90:160:31: [FAIL] [FAIL] 'XXX ','XXX ',ALICE ,BOB & !499 [FAIL] 1 [FAIL] Error: Symbol \u2018alice\u2019 at (1) has no IMPLICIT type [FAIL] /home/ubuntu/cylc-run/u-ca024/share/fcm_make/preprocess-atmos/src/um/src/atmosphere/UKCA/ukca_nmspec_mod.F90:160:36: [FAIL] [FAIL] 'XXX ','XXX ',ALICE ,BOB & !499 [FAIL] 1 [FAIL] Error: Symbol \u2018bob\u2019 at (1) has no IMPLICIT type [FAIL] compile 2.2 ! ukca_nmspec_mod.o <- um/src/atmosphere/UKCA/ukca_nmspec_mod.F90 [FAIL] ! ukca_nmspec_mod.mod : depends on failed target: ukca_nmspec_mod.o [FAIL] ! ukca_nmspec_mod.o : update task failed
You will notice that the file being pointed to is in the cylc-run/ directory:
/home/ubuntu/cylc-run/u-ca024/share/fcm_make/preprocess-atmos/src/um/src/atmosphere/UKCA/ukca_nmspec_mod.F90
To view the exact line number referenced (line 160 in this case) you should open this file printing in the job.err file and then search for the same line in the equivalent file in your working copy. You may find that the line numbers are quite different - for instance line 160 in this file in cylc-run/ directory is line 175 in the equivalent file in the working copy.
Solution to Task 4.2
You were given the task
- In UKCA, make the code changes required to include these species, ALICE and BOB, into the StratTrop chemical mechanism. You should set the conversion factor for each of these to 1.0. In the
nm_specarray you should put these into slots 498 for ALICE and 499 for BOB.
For a working Rose suite that has completed this task, please see
- vm:
u-ca024@181072
The specific Rose changes made are:
vm:
Index: app/fcm_make/rose-app.conf =================================================================== --- app/fcm_make/rose-app.conf (revision 181051) +++ app/fcm_make/rose-app.conf (revision 181072) @@ -43,4 +43,4 @@ thread_utils=false timer_version=3A um_rev=$BASE_UM_REV -um_sources=branches/dev/lukeabraham/vn11.8_UKCA_Tutorial_Solns@93557 +um_sources=branches/dev/lukeabraham/vn11.8_UKCA_Tutorial_Solns@93589
These differences can be found in the file Tutorials/vn11.8/worked_solutions/Task04.2/Task04.2_rose.patch.
The specific UM changes made are:
Index: src/atmosphere/UKCA/ukca_chem_master.F90
===================================================================
--- src/atmosphere/UKCA/ukca_chem_master.F90 (revision 93557)
+++ src/atmosphere/UKCA/ukca_chem_master.F90 (revision 93589)
@@ -94,7 +94,7 @@
INTEGER, PARAMETER :: rp = 128 ! RO2-Permutation chemistry (ST only)
! Define size of master chemistry
-INTEGER, PARAMETER :: n_chch_master = 327 ! number of known species
+INTEGER, PARAMETER :: n_chch_master = 329 ! number of known species
INTEGER, PARAMETER :: n_het_master = 18 ! number of heterogeneous reactions
INTEGER, PARAMETER :: n_dry_master = 149 ! number of dry deposition reactions
INTEGER, PARAMETER :: n_wet_master = 106 ! number of wet deposition reactions
@@ -843,7 +843,11 @@
! 270 DD:140,WD: 96,
chch_t1(270,'ARNOH17 ',1,'TR ',' ',1,1,cs,0,0,107), &
! 271 DD:141, ANHY ~ MALANHY (327)
-chch_t1(271,'ANHY ',1,'TR ',' ',1,0,cs,0,0,107) &
+chch_t1(271,'ANHY ',1,'TR ',' ',1,0,cs,0,0,107), &
+! 272
+chch_t1(272,'ALICE ',1,'TR ',' ',0,0,st,0,0,111), &
+! 273
+chch_t1(273,'BOB ',1,'TR ',' ',0,0,st,0,0,111) &
/)
! ----------------------------------------------------------------------
Index: src/atmosphere/UKCA/ukca_constants.F90
===================================================================
--- src/atmosphere/UKCA/ukca_constants.F90 (revision 93557)
+++ src/atmosphere/UKCA/ukca_constants.F90 (revision 93589)
@@ -343,6 +343,10 @@
! Same as Sec_org
REAL, PARAMETER :: c_cri = 5.1782
+! Additions for UKCA Tutorial
+REAL, PARAMETER :: C_ALICE = 1.0000
+REAL, PARAMETER :: C_BOB = 1.0000
+
! molecular masses in g/mol of emitted species,
! for budget calculations
REAL, PARAMETER :: m_ho2 = 33.007
Index: src/atmosphere/UKCA/ukca_config_defs_mod.F90
===================================================================
--- src/atmosphere/UKCA/ukca_config_defs_mod.F90 (revision 93557)
+++ src/atmosphere/UKCA/ukca_config_defs_mod.F90 (revision 93589)
@@ -315,7 +315,7 @@
(/'NO ','CH4 ','CO ','HCHO ', &
'C2H6 ','C3H8 ','Me2CO ','MeCHO ', &
'C5H8 ', nm_meoh_em ,'NO_aircrft'/)
- n_chem_tracers = 71 ! No chem tracers
+ n_chem_tracers = 73 ! No chem tracers
nr_therm = 220 ! thermal reactions
nr_phot = 55 ! photolytic (ATA)
@@ -331,7 +331,7 @@
'SO2_high ','NH3 ','DMS ','SO2_nat ', &
'BC_biomass','OC_biomass','NO_aircrft'/)
n_aero_tracers = 12
- n_chem_tracers = 71 ! No chem tracers
+ n_chem_tracers = 73 ! No chem tracers
IF (ukca_config%l_ukca_trophet) THEN
nr_therm = 241 ! thermal reactions
ELSE
Index: src/atmosphere/UKCA/ukca_nmspec_mod.F90
===================================================================
--- src/atmosphere/UKCA/ukca_nmspec_mod.F90 (revision 93557)
+++ src/atmosphere/UKCA/ukca_nmspec_mod.F90 (revision 93589)
@@ -172,7 +172,7 @@
'XXX ','XXX ','XXX ','XXX ','XXX ', & !485
'XXX ','XXX ','XXX ','XXX ','XXX ', & !490
'XXX ','XXX ','XXX ','XXX ','XXX ', & !495
-'XXX ','XXX ','XXX ','XXX ' & !499
+'XXX ','XXX ','ALICE ','BOB ' & !499
/)
IF (l_ukca_raq .OR. l_ukca_raqaero) THEN
Index: src/atmosphere/UKCA/ukca_cspecies.F90
===================================================================
--- src/atmosphere/UKCA/ukca_cspecies.F90 (revision 93557)
+++ src/atmosphere/UKCA/ukca_cspecies.F90 (revision 93589)
@@ -180,7 +180,7 @@
c_nucarb12, c_udcarb8, c_udcarb11, c_udcarb14, c_tncarb26, c_tncarb10, &
c_tncarb12, c_tncarb11, c_ccarb12, c_tncarb15, c_rcooh25, c_txcarb24, &
c_txcarb22, c_ru12pan, c_rtn26pan, c_aroh14, c_aroh17, c_arnoh14, &
- c_arnoh17, c_anhy, c_cri
+ c_arnoh17, c_anhy, c_cri, c_alice, c_bob
USE asad_mod, ONLY: advt, nadvt, speci, jpctr, jpspec
USE ukca_config_specification_mod, ONLY: ukca_config
@@ -449,6 +449,9 @@
WHERE (advt == 'RAROH17 ') c_species = c_cri
WHERE (advt == 'PASSIVE O3') c_species = 1.0
WHERE (advt == 'AGE OF AIR') c_species = 1.0
+! UKCA Tutorial species
+WHERE (advt == 'ALICE ') c_species = c_alice
+WHERE (advt == 'BOB ') c_species = c_bob
! non-advected tracers
c_na_species=0.0
These differences can be found in the file Tutorials/vn11.8/worked_solutions/Task04.2/Task04.2_code.patch.
Sample output from this task can be found at Tutorials/vn11.8/sample_output/Task04.2/atmosa.pa19810901_00.
Checklist
- ☐ If required, copy an appropriate Rose suite to use to test your changes.
- ☐ Make a new ticket on the Met Office SRS Trac pages.
- ☐ Make a branch at the required MetUM version using
fcm branch-create. - ☐ Checkout your branch using
fcm checkout. - ☐ Choose slots for your new tracer(s) from the list in
ukca_nmspec_mod.F90. - ☐ Make the required changes to branch's STASHmaster_A file, to add the new tracers.
- ☐ Add help text for your tracers in your branch's STASHmaster-meta.conf file.
- ☐
fcm committhe STASH changes to your branch. - ☐ Using a text editor, open the
app/um/rose-app.conffile from yourroses/[SUITE-ID]directory, and add the lineSTASHMASTER=STASHmasterin the[env]block, then save and close the file. - ☐ Using a text editor, open the
rose-suite.conffile from yourroses/[SUITE-ID]directory, and add the following lines to the top of the file, before saving and closing it:
[file:app/um/file/STASHmaster] source=fcm:um.xm_br/dev/[your MOSRS userid]/vnX.Y_your_branch_name/rose-meta/um-atmos/HEAD/etc/stash/STASHmaster@HEAD
- ☐ Point the metadata in your suite to the
rose-meta/um-atmos/HEADof your branch's working copy. - ☐ Include your branch in your suite at: fcm_make env Sources.
- ☐ Initialise your tracers in your suite at: um namelist Reconfiguration and Ancillary Control Configure ancils and initialise dump fields.
- ☐ Output your tracers in STASH at: um namelist Model Input and Output STASH Requests and Profiles STASH Requests.
- ☐ Run the TidyStashTransform transform macro.
- ☐ Save your suite.
- ☐ In the
roses/[SUITE-ID]directory, runfcm committo commit your changes to the repository. - ☐ Run your suite.
- ☐ Add your new tracer(s) from into the list in
ukca_nmspec_mod.F90. - ☐ Add the tracer(s) to the
chch_defs_strattroparray inukca_chem_master.F90. - ☐ Increase the value of
n_chem_tracersinukca_config_defs_mod.F90. - ☐ Add a C_species value for each tracer in
ukca_constants.F90. - ☐ Append the CASE statement in
ukca_cspecies.F90to add this value to thec_speciesarray. - ☐
fcm committhe changes to your branch. - ☐ Run your suite again.
Written by Luke Abraham 2020