Difference between revisions of "UKCA Chemistry and Aerosol UMvn13.0 Tutorial 1"
| (31 intermediate revisions by the same user not shown) | |||
| Line 7: | Line 7: | ||
| Time to Complete || '''Under 1 hour''' |
| Time to Complete || '''Under 1 hour''' |
||
|- |
|- |
||
| − | | Video instructions || |
+ | | Video instructions || [https://www.youtube.com/watch?v=n3yE-oYy-gU&t=89s Walkthrough (YouTube)] |
|} |
|} |
||
| − | '''Remember to run <tt>mosrs-cache-password</tt> |
+ | '''Remember to run <tt>mosrs-cache-password</tt>.''' |
| + | |||
| + | ==What you will learn in this tutorial== |
||
| + | |||
| + | In this tutorial you will learn how to copy a UKCA Box Model suite, run it, and plot the output. |
||
==Copying and Running an Existing Rose Suite== |
==Copying and Running an Existing Rose Suite== |
||
| Line 23: | Line 27: | ||
|} |
|} |
||
| − | + | You will need to login to the VM using SSH via a Terminal |
|
| + | ssh -X ukca |
||
| − | vagrant ssh |
||
| + | When you first login to your virtual machine you should then be asked for your Met Office Science Repository Service (MOSRS) password. Once you enter this, if this is the first time you are logging in, you will also be asked for your MOSRS username. This will likely be your name in lower case, and is '''not''' your username on the VM. |
||
| − | If you are using a VM provided for you, you should connect via X2Go or via a Terminal. |
||
| − | |||
| − | You should then be asked for your Met Office Science Repository Service (MOSRS) password. Once you enter this, if this is the first time you are logging in, you will also be asked for your MOSRS username. This will likely be your name in lower case, and is '''not''' your username on the VM. |
||
Then copy the rose suite by: |
Then copy the rose suite by: |
||
| Line 62: | Line 64: | ||
rose suite-run |
rose suite-run |
||
| − | in the terminal. |
+ | in the terminal. For this suite to run you need the UKCA branch |
| + | |||
| + | fcm:ukca.x_br/dev/lukeabraham/um13.0_ukca_box |
||
| + | |||
| + | checked-out into your <tt>home/</tt> directory. This has been done for you. |
||
If you are using your own VM, times will vary depending on the specifications of the host. It may take 2 or 3 minutes to compile the code (''fcm_make'') for the first time (and a few seconds when recompiling), followed by another few seconds to run UKCA itself (''ukca''). Any unsuccessful jobs will be highlighted in red and have a status of "failed". |
If you are using your own VM, times will vary depending on the specifications of the host. It may take 2 or 3 minutes to compile the code (''fcm_make'') for the first time (and a few seconds when recompiling), followed by another few seconds to run UKCA itself (''ukca''). Any unsuccessful jobs will be highlighted in red and have a status of "failed". |
||
| Line 70: | Line 76: | ||
==Version Control== |
==Version Control== |
||
| − | Rose suites are all held under version control, using [ |
+ | Rose suites are all held under version control, using [https://www.metoffice.gov.uk/research/weather/weather-science-it/fcm fcm]. When making changes to a suite, you will need to '''save''' it before you can '''run''' the suite. Once you are happy with the settings, you can also '''commit''' these changes back to the repository - to do this change directory to the |
/home/'''$USER'''/roses/'''[SUITE-ID]''' |
/home/'''$USER'''/roses/'''[SUITE-ID]''' |
||
| Line 105: | Line 111: | ||
!style="text-align:top;|Example |
!style="text-align:top;|Example |
||
|- |
|- |
||
| − | | Most recent ''job.out'' files || |
+ | | Most recent ''job.out'' files || <tt>/home/vagrant/cylc-run/'''[SUITE-ID]'''/log/job/1/'''[JOB NAME]'''/NN</tt> || <tt>/home/vagrant/cylc-run/u-cq782/log/job/1/ukca/NN</tt> |
|- |
|- |
||
| − | | Processor output (while running) || |
+ | | Processor output (while running) || <tt>/home/vagrant/cylc-run/'''[SUITE-ID]'''/work/1/'''[JOB NAME]'''/pe_output</tt> || <tt>/home/vagrant/cylc-run/u-cq782/work/1/ukca/pe_output</tt> |
|- |
|- |
||
| − | | UKCA Box Model output text files in ASCII format<br/> |
+ | | UKCA Box Model output text files in ASCII format<br/><tt>tracer_out.csv</tt><br/><tt>flux_out.csv</tt> || <tt>/home/vagrant/cylc-run/'''[SUITE-ID]'''/work/1/ukca</tt> || <tt>/home/vagrant/cylc-run/u-cq782/work/1/ukca</tt> |
|} |
|} |
||
| Line 115: | Line 121: | ||
* The <tt>job.out</tt> files can also be viewed through the Gcylc GUI right-click menu from each job. |
* The <tt>job.out</tt> files can also be viewed through the Gcylc GUI right-click menu from each job. |
||
| − | * UKCA Box Model output file will be <tt>tracer_out.csv</tt> for tracers and <tt>flux_out.csv</tt> for chemical reaction fluxes. These can be renamed in Rose in the '''ukca <math>\rightarrow</math> Model Input and Output''' panel. |
+ | * UKCA Box Model output file will be <tt>tracer_out.csv</tt> for tracers and <tt>flux_out.csv</tt> for chemical reaction fluxes. These can be renamed in Rose in the '''ukca <math>\rightarrow</math> namelist <math>\rightarrow</math> Model Input and Output''' panel. |
| + | |||
| + | ==Exploring Rose== |
||
| + | |||
| + | Take a look at your suite, and click through the options. There are 2 key sections that you will need to consider: |
||
| + | |||
| + | ===<tt>fcm_make</tt>=== |
||
| + | |||
| + | This section controls which fcm branches are used by the model, and also what compiler settings are used etc. You should go to '''fcm_make <math>\rightarrow</math> env <math>\rightarrow</math> Sources''' to see what branches are being used. During the course of these tutorials you will make a branch and add it here to complete the tutorial tasks. You can add both relative paths to the repository (starting with <tt>branches/</tt>) and absolute paths to a working copy (starting with <tt>/home/</tt>). |
||
| + | |||
| + | ===<tt>ukca</tt>=== |
||
| + | |||
| + | Click the arrow by '''ukca''' to view the list of sections. We'll be most interested in the '''namelist''' section, which contains all the settings that configure the science that can be done by UKCA. Take a look through the panels and sub-panels in this section to get a feel for how it is organised. |
||
==Viewing Output== |
==Viewing Output== |
||
| − | === |
+ | ===gnuplot=== |
| + | <gallery widths=400px heights=350px> |
||
| + | Image:T01_gnuplot.png|Plotting O3 concentrations using Gnuplot |
||
| + | Image:T01_o3_002.png|O3 mass-mixing ratio plotted using gnuplot |
||
| + | </gallery> |
||
| + | The quickest and easiest way to plot UKCA box model output is using '''[http://www.gnuplot.info/ Gnuplot]''', which is a relatively simple command-line plotting tool. The box model outputs tracer concentrations and chemical reaction fluxes every timestep to the files '''<tt>tracer_out.csv</tt>''' and '''<tt>flux_out.csv</tt>''' respectively. |
||
| − | ===Met Office Unified Model=== |
||
| + | To plot these files, change into your suite's cylc-run directory and navigate to the ukca directory in work, e.g. |
||
| − | [[Image:Vn10.9_base_output.png|thumb|300px|right|Example output from the UKCA training suite.]] |
||
| + | cd /home/vagrant/cylc-run/u-cq782/work/1/ukca |
||
| − | For more detailed plotting, tools such as the [http://scitools.org.uk/iris/ Iris] and [http://cfpython.bitbucket.org/ cf-python] libraries can be used to view UM file formats directly. However, for quick viewing, [http://cms.ncas.ac.uk/documents/xconv/ Xconv] is a very useful tool. Information on how to install Xconv on the VM can be found [[UKCA_Chemistry_and_Aerosol_Tutorials:_Things_to_know_before_you_start_the_vn11.8_practicals#Xconv|here]]. |
||
| + | then load gnuplot by typing |
||
| − | To view these files, do |
||
| + | gnuplot |
||
| − | '''xconv atmosa.pa19810901_00''' |
||
| + | To plot the tracer file, type |
||
| − | As well as viewing files, you can use Xconv to convert these files to netCDF, by filling in the '''''Output file name:''''' box (e.g. ''foo.nc''), and then clicking '''convert'''. If no path is defined, this will save the file in the same directory that you opened Xconv from. |
||
| + | plot "tracer_out.csv" using 1:3 with linespoints |
||
| − | Example output from the UKCA training suite can be found at |
||
| + | |||
| + | which can be shortened to |
||
| + | |||
| + | p "tracer_out.csv" u 1:3 w lp |
||
| + | |||
| + | The file can be saved from the export menu on the plot itself. Additional options for the [http://gnuplot.info/docs_5.5/loc9418.html <tt>set</tt> command] can be used to define labels, titles, change how the axis labels look, e.g. |
||
| + | |||
| + | gnuplot> set title "O3 MMR" |
||
| + | gnuplot> set xlabel "timestep number" |
||
| + | gnuplot> set ylabel "mass mixing ratio /kg kg^{-1}" |
||
| + | gnuplot> set format y "%.2t*10^{%+03T}" |
||
| + | gnuplot> unset key |
||
| + | gnuplot> p 'tracer_out.csv' u 1:3 w lp |
||
| + | |||
| + | The ordering of the species in the <tt>tracer_out.csv</tt> can be seen by opening it in e.g. a simple spreadsheet program like '''[http://www.gnumeric.org/ Gnumeric]'''. You may need to install this first using |
||
| + | |||
| + | sudo apt install gnumeric |
||
| + | |||
| + | before opening the file |
||
| + | |||
| + | gnumeric tracer_out.csv |
||
| + | |||
| + | The column number is given in the first line with the tracer name in the line below. You could also open the file in <tt>emacs</tt> and then <tt>M-x toggle-truncate-lines</tt>. An example of this header for the StratTrop chemistry with GLOMAP-mode aerosols is |
||
| + | |||
| + | <pre style="white-space: pre;"> |
||
| + | # COL 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96 |
||
| + | # Timestep, O(3P), O3, N, NO, NO3, NO2, N2O5, HO2NO2, HONO2, H2O2, CH4, CO, HCHO, MeOOH, H, H2O, OH, HO2, Cl, Cl2O2, ClO, OClO, Br, BrO, BrCl, BrONO2, N2O, HCl, HOCl, HBr, HOBr, ClONO2, CFCl3, CF2Cl2, MeBr, HONO, C2H6, EtOOH, MeCHO, PAN, C3H8, n-PrOOH, i-PrOOH, EtCHO, Me2CO, MeCOCH2OOH, PPAN, MeONO2, C5H8, ISOOH, ISON, MACR, MACROOH, MPAN, HACET, MGLY, NALD, HCOOH, MeCO3H, MeCO2H, H2, MeOH, DMS, SO2, H2SO4, MSA, DMSO, NH3, CS2, COS, H2S, SO3, Monoterp, Sec_Org, Nuc_SOL_N, Nuc_SOL_SU, Ait_SOL_N, Ait_SOL_SU, Ait_SOL_BC, Ait_SOL_OM, Acc_SOL_N, Acc_SOL_SU, Acc_SOL_BC, Acc_SOL_OM, Acc_SOL_SS, Cor_SOL_N, Cor_SOL_SU, Cor_SOL_BC, Cor_SOL_OM, Cor_SOL_SS, Ait_INS_N, Ait_INS_BC, Ait_INS_OM, Nuc_SOL_OM, PASSIVE O3 |
||
| + | </pre> |
||
| + | |||
| + | The <tt>flux_out.csv</tt> file has a more complicated header as it also gives all the chemical reactants and products, e.g. |
||
| + | |||
| + | <pre style="white-space: pre;"> |
||
| + | # Timestep, UKCA reaction fluxes (units = molecules.cm^-3.s^-1) |
||
| + | # COL 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210, 211, 212, 213, 214, 215, 216, 217, 218, 219, 220, 221, 222, 223, 224, 225, 226, 227, 228, 229, 230, 231, 232, 233, 234, 235, 236, 237, 238, 239, 240, 241, 242, 243, 244, 245, 246, 247, 248, 249, 250, 251, 252, 253, 254, 255, 256, 257, 258, 259, 260, 261, 262, 263, 264, 265, 266, 267, 268, 269, 270, 271, 272, 273, 274, 275, 276, 277, 278, 279, 280, 281, 282, 283, 284, 285, 286, 287, 288, 289, 290, 291, 292, 293, 294, 295, 296, 297, 298, 299, 300, 301, 302, 303, 304, 305, 306, 307, 308, 309, 310 |
||
| + | # PROD 1, Cl2O2, HO2NO2, PAN, PPAN, MPAN, N2O5, EtOOH, H2O2, HCHO, HCHO, HO2NO2, HONO2, MeCHO, MeCHO, MeOOH, N2O5, NO2, NO3, NO3, O2, O3, O3, PAN, HONO, EtCHO, Me2CO, n-PrOOH, i-PrOOH, MeCOCH2OOH, PPAN, MeONO2, ISOOH, ISON, MACR, MPAN, MACROOH, MACROOH, HACET, MGLY, NALD, MeCO3H, BrCl, BrO, BrONO2, BrONO2, O2, OClO, NO, HOBr, N2O, H2O, ClONO2, ClONO2, HCl, HOCl, Cl2O2, CFCl3, CF2Cl2, MeBr, CH4, CO2, HO2NO2, CS2, COS, H2SO4, SO3, Br, Br, Br, Br, Br, BrO, BrO, BrO, BrO, BrO, BrO, BrO, CF2Cl2, CFCl3, Cl, Cl, Cl, Cl, Cl, Cl, Cl, Cl, Cl, Cl, Cl, Cl, Cl, ClO, ClO, ClO, ClO, ClO, ClO, ClO, EtCO3, EtCO3, EtOO, EtOO, H, H, H, H, H, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, MeBr, MeBr, MeBr, MeCO3, MeCO3, MeCOCH2OO, MeCOCH2OO, MeOO, MeOO, MeOO, N, N, N, N2O5, NO, NO, NO, NO, NO, NO, NO2, NO2, NO3, NO3, NO3, NO3, NO3, NO3, NO3, O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O3, O3, O3, O3, O3, O3, O3, OClO, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, i-PrOO, i-PrOO, n-PrOO, n-PrOO, CS2, CS2, DMS, DMS, DMSO, H2S, H2S, COS, COS, SO2, SO3, Monoterp, Monoterp, Monoterp, MeOO, MeOO, EtOO, EtOO, n-PrOO, n-PrOO, i-PrOO, i-PrOO, MeCO3, EtCO3, MeCOCH2OO, MeCOCH2OO, ISO2, MACRO2, MACRO2, HO2, O(3P), O(3P), O(3P), O(1D), BrO, ClO, ClO, H, HO2, HO2, OH, OH, OH, MeCO3, EtCO3, MACRO2, NO2, NO, SO2, DMS, DMS, ClONO2, ClONO2, HOCl, N2O5, N2O5, SO2, SO2, SO2 |
||
| + | # PROD 2, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , Cl2O2, HCHO, HO2, O3, OClO, BrO, ClO, ClO, ClO, HO2, NO, OH, O(1D), O(1D), CH4, Cl2O2, ClONO2, H2, H2O2, HCHO, HO2, HO2, HOCl, MeOOH, NO3, O3, OClO, ClO, ClO, ClO, HO2, MeOO, NO, NO3, NO, NO3, NO, NO3, HO2, HO2, HO2, NO2, O3, EtCO3, EtCO3, EtOO, HO2, ISO2, MACRO2, MeCO3, MeCO3, MeCO3, MeCOCH2OO, MeOO, MeOO, NO, NO3, O3, i-PrOO, n-PrOO, Cl, O(1D), OH, NO, NO3, NO, NO3, NO, NO, NO3, NO, NO2, O2, H2O, ISO2, ISO2, MACRO2, MACRO2, NO3, O3, NO3, O3, Br, C5H8, EtCHO, HCHO, MGLY, Me2CO, MeCHO, CH4, CH4, CH4, CO2, H2, H2O, HBr, HBr, HCl, HCl, HCl, N2, N2O, N2O, O2, O3, O3, BrO, ClO, ClONO2, H2O2, HBr, HCl, HO2, HOCl, NO2, NO3, O3, OClO, OH, C5H8, C5H8, C5H8, MACR, MACR, MACR, MACR, NO, C2H6, C3H8, C3H8, C5H8, CH4, CO, ClO, ClO, ClONO2, EtCHO, EtOOH, EtOOH, H2, H2O2, HACET, HBr, HCHO, HCOOH, HCl, HO2, HO2NO2, HOCl, HONO, HONO2, ISON, ISOOH, MACR, MACR, MACROOH, MGLY, MPAN, Me2CO, Me2CO, MeCHO, MeCO2H, MeCO3H, MeCOCH2OOH, MeCOCH2OOH, MeOH, MeONO2, MeOOH, MeOOH, NALD, NO3, O3, OClO, OH, PAN, PPAN, i-PrOOH, i-PrOOH, n-PrOOH, n-PrOOH, NO, NO3, NO, NO3, O(3P), OH, OH, NO3, OH, O(3P), OH, O(3P), OH, O3, H2O, OH, O3, NO3, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, EtCO3, O2, NO, NO2, N2, NO2, ClO, NO2, O2, HO2, NO2, NO, NO2, OH, NO2, NO2, NO2, NO3, NO, OH, OH, OH, H2O, HCl, HCl, H2O, HCl, H2O2, O3, O3 |
||
| + | # REACT 1, ClO, NO2, MeCO3, EtCO3, MACRO2, NO2, MeCHO, OH, HO2, H2, HO2, OH, MeOO, CH4, HO2, NO3, NO, NO, NO2, O(3P), O2, O2, MeCO3, OH, EtOO, MeCO3, EtCHO, Me2CO, MeCO3, EtCO3, HO2, OH, NO2, MeCO3, MACRO2, OH, HACET, MeCO3, MeCO3, HCHO, MeOO, Br, Br, Br, BrO, O(3P), O(3P), N, OH, N2, OH, Cl, ClO, H, OH, Cl, Cl, Cl, Br, MeOO, CO, OH, COS, CO, SO3, SO2, BrCl, HBr, HBr, BrO, BrO, Br, Br, Br, BrCl, HOBr, Br, Br, Cl, Cl, HCl, Cl, Cl, HCl, HCl, HCl, ClO, HCl, Cl, HCl, ClO, ClO, ClO, Cl, Cl, Cl, HOCl, Cl, Cl, Cl, EtOO, EtOO, MeCHO, MeCHO, H2, O(3P), OH, OH, OH, O2, O3, EtOOH, H2O2, ISOOH, MACROOH, MeCO2H, MeCO3H, OH, MeCOCH2OOH, HCHO, MeOOH, OH, OH, OH, i-PrOOH, n-PrOOH, Br, Br, Br, MeOO, MeOO, MeCO3, MeCO3, HO2, MeONO2, HO2, N2, N2O, NO, HONO2, ISON, NO2, MGLY, NO2, NO2, NO2, NO, NO3, BrO, ISON, HONO2, HONO2, MeCO3, HONO2, HONO2, HCHO, HCHO, OH, O(3P), OH, OH, HBr, OH, H, O(3P), OH, O(3P), N2, NO, O(3P), O2, O2, O2, Cl, ClO, OH, OH, OH, OH, OH, NO, O2, O2, O2, O2, HO2, MACR, MeOO, MGLY, MGLY, OH, OH, NO2, H2O, i-PrOO, n-PrOO, ISO2, H2O, H, HCl, HO2, HOCl, H2O, H2O, H2O, H2O, H2O, MGLY, H2O, H2O, CO2, H2O, H2O, H2O, ClO, H2O, H2O, HACET, MACR, MACRO2, MACRO2, MACRO2, MeCO3, HACET, H2O, H2O, H2O, MeOO, MeCO3, H2O, OH, HO2, HCHO, H2O, H2O, HCHO, HO2, HO2, HOCl, H2O, HCHO, MeCHO, Me2CO, i-PrOO, EtCHO, n-PrOO, Me2CO, Me2CO, EtCHO, EtCHO, CO, COS, SO2, SO2, SO2, OH, SO2, CO, CO2, SO3, H2SO4, Sec_Org, Sec_Org, Sec_Org, HO2, MeOH, HO2, MeCHO, HO2, EtCHO, HO2, Me2CO, MeCO2H, , HCHO, HACET, MACR, HACET, HO2, OH, O3, NO2, NO3, N2O, BrONO2, Cl2O2, ClONO2, HO2, H2O2, HO2NO2, HONO, HONO2, H2O2, PAN, PPAN, MPAN, N2O5, NO2, SO3, DMSO, SO2, HOCl, Cl, Cl, HONO2, Cl, , , |
||
| + | # REACT 2, ClO, HO2, NO2, NO2, NO2, NO3, HO2, OH, HO2, CO, NO2, NO2, HO2, CO, HCHO, NO2, O(3P), O2, O(3P), O(3P), O(1D), O(3P), NO2, NO, HO2, MeOO, HO2, HO2, HCHO, NO2, HCHO, MACR, MACR, HCHO, NO2, HO2, CO, HCHO, CO, CO, OH, Cl, O(3P), NO3, NO2, O(1D), ClO, O(3P), Br, O(1D), H, NO3, NO2, Cl, Cl, Cl, Cl, Cl, H, H, O(3P), NO3, SO2, SO2, OH, O(3P), Cl, CO, O2, O2, ClO, Br, Cl, OClO, O2, O2, NO2, HO2, ClO, Cl, MeOO, Cl, Cl, H, HO2, CO, OH, O2, Cl, MeOO, NO2, O2, ClO, Cl, Cl, OClO, O2, HCHO, NO2, O2, CO2, CO2, HO2, HO2, O2, H2O, OH, NO, O2, , , , , , , O3, , MeOO, , , , NO2, NO2, O2, , , HCl, OH, H2O, CO2, CO2, HCHO, HCHO, HCHO, , HCHO, O(3P), O(3P), O(3P), HONO2, , MACR, HCHO, MeCO3, NO2, , NO2, , NO2, , EtCO3, HO2, CO, MeCOCH2OO, MeCO3, H2, HO2, MeOO, CO2, H, OH, O(3P), Br, ClO, HCl, Cl, N2, O2, NO, O2, O(3P), O2, Br, O2, NO3, HO2, Br, Cl, O2, ClO, O2, NO2, O2, ClO, H, OH, HCHO, HCOOH, HCOOH, HCOOH, MeCO3, MeCO3, ClO, EtOO, H2O, H2O, , MeOO, CO2, O2, Cl, NO3, EtCO3, EtOO, MeCHO, H, HO2, HO2, Br, HO2, H2O, Cl, , NO2, H2O, NO2, NO3, NALD, OH, , , , CO, NO2, MeCOCH2OO, MeCOCH2OO, MeCO3, , , MeCOCH2OO, MGLY, HCHO, NO2, HCHO, MeOO, CO, NO2, O2, O2, O(3P), NO2, NO2, OH, H2O, H2O, H2O, HO2, HO2, HO2, HO2, SO2, SO2, , , MSA, SO2, H2O, SO2, SO2, , H2O, , , , HCHO, HCHO, MeCHO, , EtCHO, , Me2CO, , MeOO, EtOO, MeCO3, MGLY, HCHO, MGLY, , EtOO, , , , , , , , , O2, , , , , , , , , NO2, HO2, HO2, MeOO, HONO2, Cl, Cl, HONO2, NO2, , , |
||
| + | # REACT 3, , , , , , , OH, , CO, , , , CO, , OH, , , , , , , , , , CO, , OH, OH, OH, , NO2, HCHO, HCHO, CO, , OH, MGLY, HO2, HO2, NO2, , , , , , , , , , , , , , , , O2, Cl, , , , , , , , H, , O2, HO2, , , , O2, O2, , , , , , , ClO, , Cl, NO3, , , HO2, , , OH, , , , , O2, O2, , , HO2, , NO2, NO2, NO2, NO2, NO2, , , , , , , , , , , , , , , , , , , , , , , , , , NO2, NO2, NO2, NO2, NO2, , NO2, , , , , , HCHO, HO2, HACET, , , O2, , , , , CO, HONO2, , , , HO2, , , , , , , , , , , , , , O(3P), , , , , , , , , , , , , , , , MACRO2, CO, HO2, HO2, , , , , , , , , , , , , , , OH, , , , , CO, H, , , O2, , , , , , , , , , , , , , , , , , , H2O, OH, , NO2, , , , , H2O, H2O, , , OH, , NO2, NO2, NO2, NO2, SO2, , , , , OH, , , , , , , , , , , , , , , , , , , , , HO2, HCHO, , CO2, , , , , , , , , , , , , , , , , , , , , , , HONO2, H2O, , HONO2, , , |
||
| + | # REACT 4, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , HO2, HO2, HO2, , HO2, HCHO, , , HO2, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , HO2, , CO, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , MeCO3, H2O2, CO, CO, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , O(3P), , , , , , , , , , , , , , , , , , , , , , CO, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , |
||
| + | </pre> |
||
| + | |||
| + | ===Jupyter Notebook=== |
||
| + | |||
| + | <gallery widths=400px heights=350px> |
||
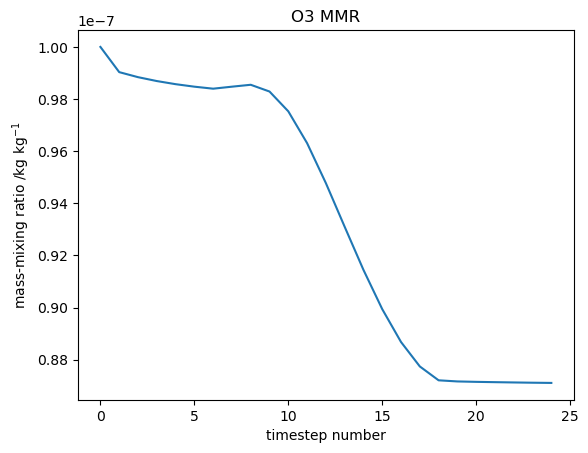
| + | Image:T01_o3_001.png|Ozone mass-mixing ration |
||
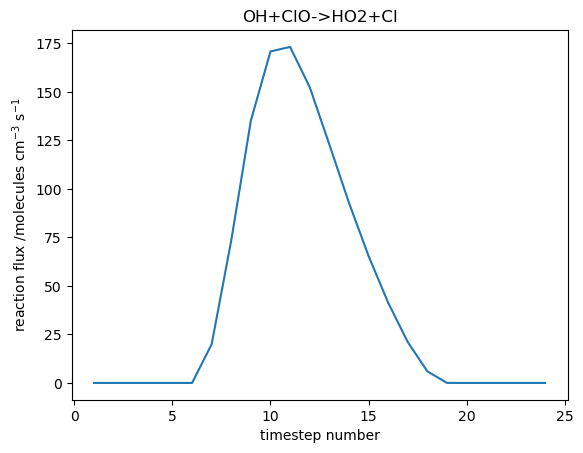
| + | Image:T01_f200_001.png|Flux through the OH+ClO->HO2+Cl reaction |
||
| + | </gallery> |
||
| + | |||
| + | An example Jupyter Notebook which plots both tracers and chemical reaction fluxes can be found in |
||
| + | |||
| + | Tutorials/UMvn13.0/notebooks/BoxModel_T01-03.ipynb |
||
| + | |||
| + | and can be viewed online here |
||
| + | |||
| + | * [https://github.com/theabro/ukca/blob/master/Tutorials/UMvn13.0/notebooks/BoxModel_T01-03.ipynb BoxModel_T01-03.ipynb] |
||
| + | |||
| + | |||
| + | You can try changing the chemical species and reaction flux selected. |
||
| + | |||
| + | Connect to the notebook using |
||
| + | |||
| + | * <div style="font-size: 140%;">'''http://localhost:4801/?token=UKCATraining'''</div> |
||
==Checklist== |
==Checklist== |
||
| Line 141: | Line 226: | ||
: <span style="font-size:20px">☐</span> Copy suites using the right-click menu |
: <span style="font-size:20px">☐</span> Copy suites using the right-click menu |
||
: <span style="font-size:20px">☐</span> Run suites using the play button |
: <span style="font-size:20px">☐</span> Run suites using the play button |
||
| + | : <span style="font-size:20px">☐</span> Plot output using gnuplot or python |
||
| + | |||
| + | [[UKCA Chemistry and Aerosol UMvn13.0 Tutorial 2|Tutorial 2]] |
||
| − | [[UKCA Chemistry and Aerosol |
+ | [[UKCA Chemistry and Aerosol Tutorials at UMvn13.0]] |
---- |
---- |
||
''Written by [[User:Nla27 | Luke Abraham]] 2022'' |
''Written by [[User:Nla27 | Luke Abraham]] 2022'' |
||
Latest revision as of 14:17, 29 January 2024
UKCA Chemistry and Aerosol Tutorials at UMvn13.0
| Difficulty | EASY |
| Time to Complete | Under 1 hour |
| Video instructions | Walkthrough (YouTube) |
Remember to run mosrs-cache-password.
What you will learn in this tutorial
In this tutorial you will learn how to copy a UKCA Box Model suite, run it, and plot the output.
Copying and Running an Existing Rose Suite
| Machine | UM Version/Configuration | Suite ID |
|---|---|---|
| vm | UMvn13.0 UKCA Box Model | u-cq774 |
You will need to login to the VM using SSH via a Terminal
ssh -X ukca
When you first login to your virtual machine you should then be asked for your Met Office Science Repository Service (MOSRS) password. Once you enter this, if this is the first time you are logging in, you will also be asked for your MOSRS username. This will likely be your name in lower case, and is not your username on the VM.
Then copy the rose suite by:
rosie copy u-cq774
This will open an editor containing text similar to this:
description=Copy of u-cq774/trunk@236265 owner=lukeabraham project=ukca title=UMvn13.0 UKCA Box Model for UKCA Tutorials # Make changes ABOVE these lines. # The "owner", "project" and "title" fields are compulsory. # Any KEY=VALUE pairs can be added. Known fields include: # "access-list", "description" and "sub-project".
You can edit the title to make it more descriptive if you like. Then save (e.g. Ctrl-s) and close the file (e.g. Ctrl-q). The press y on the keyboard when prompted and press return.
The suite will now copy and checkout to your /home/$USER/roses directory, into a sub-directory that is the same as the SUITE-ID. You should change into this directory now using cd, e.g.
cd roses/u-ab123
then open the suite by typing
rose edit &
and pressing return. You can either run the suite by pressing the play button that looks like a triangle to the top right of the options bar, or by typing
rose suite-run
in the terminal. For this suite to run you need the UKCA branch
fcm:ukca.x_br/dev/lukeabraham/um13.0_ukca_box
checked-out into your home/ directory. This has been done for you.
If you are using your own VM, times will vary depending on the specifications of the host. It may take 2 or 3 minutes to compile the code (fcm_make) for the first time (and a few seconds when recompiling), followed by another few seconds to run UKCA itself (ukca). Any unsuccessful jobs will be highlighted in red and have a status of "failed".
When the suite has finished successfully it will then become blank with the message stopped with 'succeeded' in the bottom-left corner.
Version Control
Rose suites are all held under version control, using fcm. When making changes to a suite, you will need to save it before you can run the suite. Once you are happy with the settings, you can also commit these changes back to the repository - to do this change directory to the
/home/$USER/roses/[SUITE-ID]
and then type
fcm commit
a text editor will then open, and you should type a short message describing what the changes you have made do. You should then close the editor and type y in the terminal. It is recommended that you commit frequently (even on configurations that aren't working) as this protects you against mistakes and accidental deletions etc.
These suites can be viewed on the SRS here: https://code.metoffice.gov.uk/trac/roses-u (password required)
It is recommended that you commit your suites regularly.
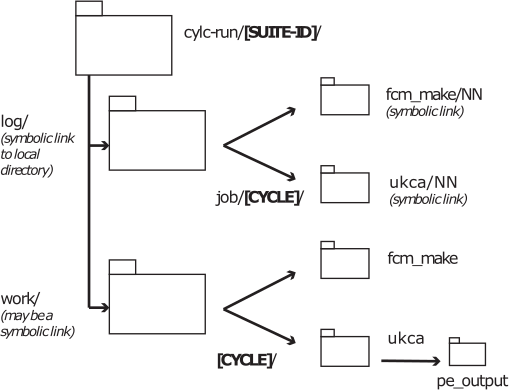
Output Directory Structure
The output directory structure of rose suites are rather complex. The schematic gives a broad overview of the general structure. Everything can be found within the [SUITE-ID] (e.g. u-cq782 etc.) directory, which can be found within your $HOME/cylc-run directory.
Within this directory there are several directories, including:
- log: a symbolic link to a directory called something like log.20220914T092709Z, which contains all the job.out (containing output from UM WRITE statements from the umPrint subroutine) and job.err files etc., as well as the script used to run the job.
- work: this contains the directories used when the job actually runs. Real-time output will be sent to files here (held in a pe_output directory. Sometimes model output will also be here.
Within these directories there will be many sub-directories. Some of these will be named from the cycle-point (labelled [CYCLE] in the graphic. For the UKCA Training Suite this will be 1. The sub-directories will (eventually) be named after the app (labelled by [JOB NAME] in the table below) that the output is from, e.g. fcm_make, ukca etc.
| Files | General Path | Example |
|---|---|---|
| Most recent job.out files | /home/vagrant/cylc-run/[SUITE-ID]/log/job/1/[JOB NAME]/NN | /home/vagrant/cylc-run/u-cq782/log/job/1/ukca/NN |
| Processor output (while running) | /home/vagrant/cylc-run/[SUITE-ID]/work/1/[JOB NAME]/pe_output | /home/vagrant/cylc-run/u-cq782/work/1/ukca/pe_output |
| UKCA Box Model output text files in ASCII format tracer_out.csv flux_out.csv |
/home/vagrant/cylc-run/[SUITE-ID]/work/1/ukca | /home/vagrant/cylc-run/u-cq782/work/1/ukca |
Notes
- The job.out files can also be viewed through the Gcylc GUI right-click menu from each job.
- UKCA Box Model output file will be tracer_out.csv for tracers and flux_out.csv for chemical reaction fluxes. These can be renamed in Rose in the ukca namelist Model Input and Output panel.
Exploring Rose
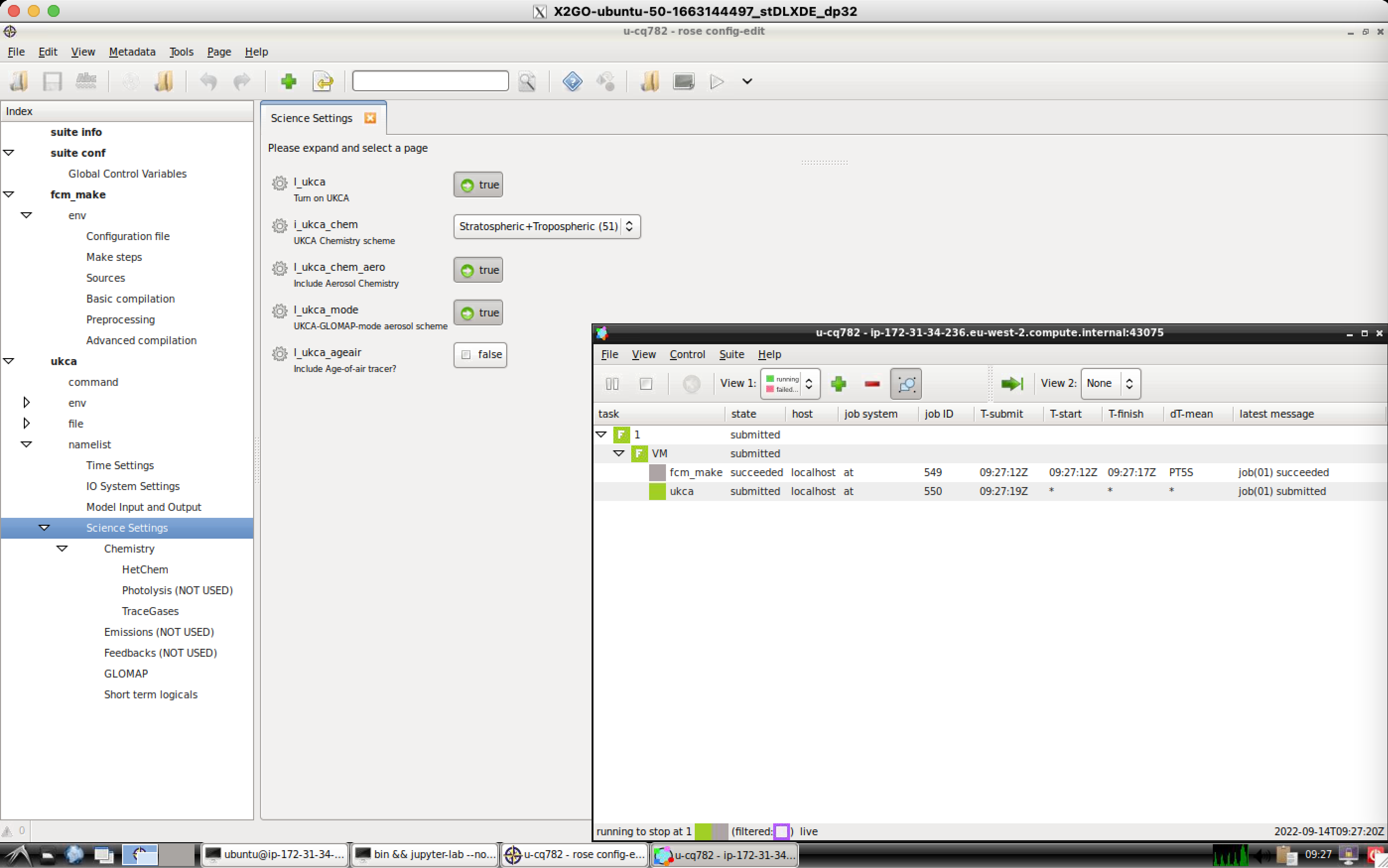
Take a look at your suite, and click through the options. There are 2 key sections that you will need to consider:
fcm_make
This section controls which fcm branches are used by the model, and also what compiler settings are used etc. You should go to fcm_make env Sources to see what branches are being used. During the course of these tutorials you will make a branch and add it here to complete the tutorial tasks. You can add both relative paths to the repository (starting with branches/) and absolute paths to a working copy (starting with /home/).
ukca
Click the arrow by ukca to view the list of sections. We'll be most interested in the namelist section, which contains all the settings that configure the science that can be done by UKCA. Take a look through the panels and sub-panels in this section to get a feel for how it is organised.
Viewing Output
gnuplot
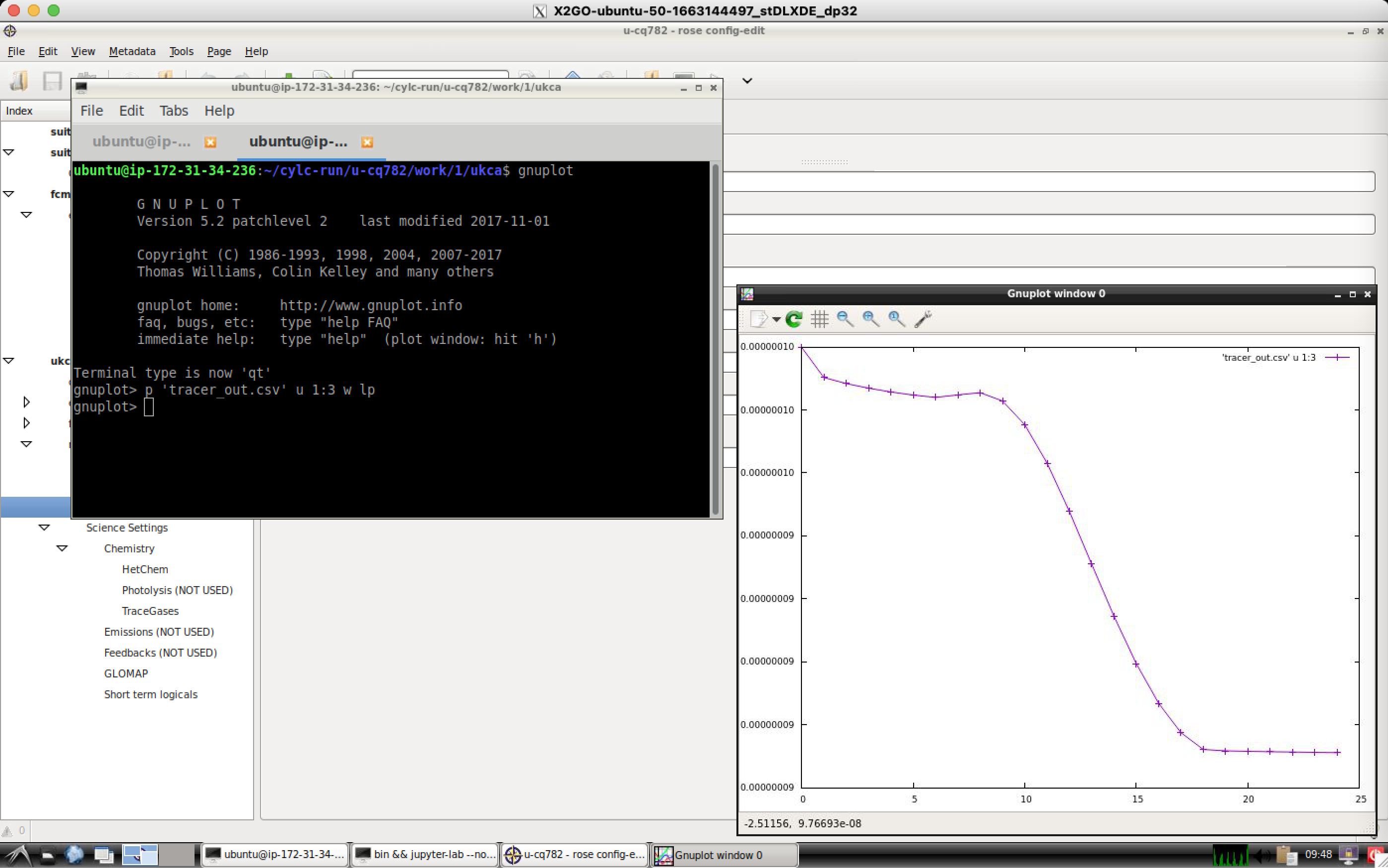
The quickest and easiest way to plot UKCA box model output is using Gnuplot, which is a relatively simple command-line plotting tool. The box model outputs tracer concentrations and chemical reaction fluxes every timestep to the files tracer_out.csv and flux_out.csv respectively.
To plot these files, change into your suite's cylc-run directory and navigate to the ukca directory in work, e.g.
cd /home/vagrant/cylc-run/u-cq782/work/1/ukca
then load gnuplot by typing
gnuplot
To plot the tracer file, type
plot "tracer_out.csv" using 1:3 with linespoints
which can be shortened to
p "tracer_out.csv" u 1:3 w lp
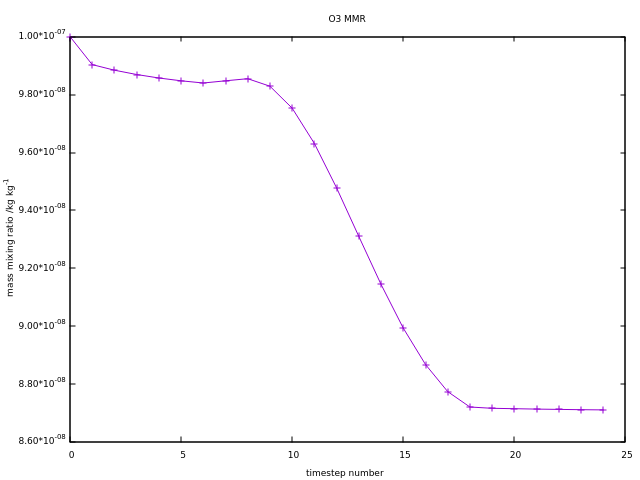
The file can be saved from the export menu on the plot itself. Additional options for the set command can be used to define labels, titles, change how the axis labels look, e.g.
gnuplot> set title "O3 MMR"
gnuplot> set xlabel "timestep number"
gnuplot> set ylabel "mass mixing ratio /kg kg^{-1}"
gnuplot> set format y "%.2t*10^{%+03T}"
gnuplot> unset key
gnuplot> p 'tracer_out.csv' u 1:3 w lp
The ordering of the species in the tracer_out.csv can be seen by opening it in e.g. a simple spreadsheet program like Gnumeric. You may need to install this first using
sudo apt install gnumeric
before opening the file
gnumeric tracer_out.csv
The column number is given in the first line with the tracer name in the line below. You could also open the file in emacs and then M-x toggle-truncate-lines. An example of this header for the StratTrop chemistry with GLOMAP-mode aerosols is
# COL 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96 # Timestep, O(3P), O3, N, NO, NO3, NO2, N2O5, HO2NO2, HONO2, H2O2, CH4, CO, HCHO, MeOOH, H, H2O, OH, HO2, Cl, Cl2O2, ClO, OClO, Br, BrO, BrCl, BrONO2, N2O, HCl, HOCl, HBr, HOBr, ClONO2, CFCl3, CF2Cl2, MeBr, HONO, C2H6, EtOOH, MeCHO, PAN, C3H8, n-PrOOH, i-PrOOH, EtCHO, Me2CO, MeCOCH2OOH, PPAN, MeONO2, C5H8, ISOOH, ISON, MACR, MACROOH, MPAN, HACET, MGLY, NALD, HCOOH, MeCO3H, MeCO2H, H2, MeOH, DMS, SO2, H2SO4, MSA, DMSO, NH3, CS2, COS, H2S, SO3, Monoterp, Sec_Org, Nuc_SOL_N, Nuc_SOL_SU, Ait_SOL_N, Ait_SOL_SU, Ait_SOL_BC, Ait_SOL_OM, Acc_SOL_N, Acc_SOL_SU, Acc_SOL_BC, Acc_SOL_OM, Acc_SOL_SS, Cor_SOL_N, Cor_SOL_SU, Cor_SOL_BC, Cor_SOL_OM, Cor_SOL_SS, Ait_INS_N, Ait_INS_BC, Ait_INS_OM, Nuc_SOL_OM, PASSIVE O3
The flux_out.csv file has a more complicated header as it also gives all the chemical reactants and products, e.g.
# Timestep, UKCA reaction fluxes (units = molecules.cm^-3.s^-1) # COL 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210, 211, 212, 213, 214, 215, 216, 217, 218, 219, 220, 221, 222, 223, 224, 225, 226, 227, 228, 229, 230, 231, 232, 233, 234, 235, 236, 237, 238, 239, 240, 241, 242, 243, 244, 245, 246, 247, 248, 249, 250, 251, 252, 253, 254, 255, 256, 257, 258, 259, 260, 261, 262, 263, 264, 265, 266, 267, 268, 269, 270, 271, 272, 273, 274, 275, 276, 277, 278, 279, 280, 281, 282, 283, 284, 285, 286, 287, 288, 289, 290, 291, 292, 293, 294, 295, 296, 297, 298, 299, 300, 301, 302, 303, 304, 305, 306, 307, 308, 309, 310 # PROD 1, Cl2O2, HO2NO2, PAN, PPAN, MPAN, N2O5, EtOOH, H2O2, HCHO, HCHO, HO2NO2, HONO2, MeCHO, MeCHO, MeOOH, N2O5, NO2, NO3, NO3, O2, O3, O3, PAN, HONO, EtCHO, Me2CO, n-PrOOH, i-PrOOH, MeCOCH2OOH, PPAN, MeONO2, ISOOH, ISON, MACR, MPAN, MACROOH, MACROOH, HACET, MGLY, NALD, MeCO3H, BrCl, BrO, BrONO2, BrONO2, O2, OClO, NO, HOBr, N2O, H2O, ClONO2, ClONO2, HCl, HOCl, Cl2O2, CFCl3, CF2Cl2, MeBr, CH4, CO2, HO2NO2, CS2, COS, H2SO4, SO3, Br, Br, Br, Br, Br, BrO, BrO, BrO, BrO, BrO, BrO, BrO, CF2Cl2, CFCl3, Cl, Cl, Cl, Cl, Cl, Cl, Cl, Cl, Cl, Cl, Cl, Cl, Cl, ClO, ClO, ClO, ClO, ClO, ClO, ClO, EtCO3, EtCO3, EtOO, EtOO, H, H, H, H, H, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, HO2, MeBr, MeBr, MeBr, MeCO3, MeCO3, MeCOCH2OO, MeCOCH2OO, MeOO, MeOO, MeOO, N, N, N, N2O5, NO, NO, NO, NO, NO, NO, NO2, NO2, NO3, NO3, NO3, NO3, NO3, NO3, NO3, O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(1D), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O(3P), O3, O3, O3, O3, O3, O3, O3, OClO, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, OH, i-PrOO, i-PrOO, n-PrOO, n-PrOO, CS2, CS2, DMS, DMS, DMSO, H2S, H2S, COS, COS, SO2, SO3, Monoterp, Monoterp, Monoterp, MeOO, MeOO, EtOO, EtOO, n-PrOO, n-PrOO, i-PrOO, i-PrOO, MeCO3, EtCO3, MeCOCH2OO, MeCOCH2OO, ISO2, MACRO2, MACRO2, HO2, O(3P), O(3P), O(3P), O(1D), BrO, ClO, ClO, H, HO2, HO2, OH, OH, OH, MeCO3, EtCO3, MACRO2, NO2, NO, SO2, DMS, DMS, ClONO2, ClONO2, HOCl, N2O5, N2O5, SO2, SO2, SO2 # PROD 2, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , Cl2O2, HCHO, HO2, O3, OClO, BrO, ClO, ClO, ClO, HO2, NO, OH, O(1D), O(1D), CH4, Cl2O2, ClONO2, H2, H2O2, HCHO, HO2, HO2, HOCl, MeOOH, NO3, O3, OClO, ClO, ClO, ClO, HO2, MeOO, NO, NO3, NO, NO3, NO, NO3, HO2, HO2, HO2, NO2, O3, EtCO3, EtCO3, EtOO, HO2, ISO2, MACRO2, MeCO3, MeCO3, MeCO3, MeCOCH2OO, MeOO, MeOO, NO, NO3, O3, i-PrOO, n-PrOO, Cl, O(1D), OH, NO, NO3, NO, NO3, NO, NO, NO3, NO, NO2, O2, H2O, ISO2, ISO2, MACRO2, MACRO2, NO3, O3, NO3, O3, Br, C5H8, EtCHO, HCHO, MGLY, Me2CO, MeCHO, CH4, CH4, CH4, CO2, H2, H2O, HBr, HBr, HCl, HCl, HCl, N2, N2O, N2O, O2, O3, O3, BrO, ClO, ClONO2, H2O2, HBr, HCl, HO2, HOCl, NO2, NO3, O3, OClO, OH, C5H8, C5H8, C5H8, MACR, MACR, MACR, MACR, NO, C2H6, C3H8, C3H8, C5H8, CH4, CO, ClO, ClO, ClONO2, EtCHO, EtOOH, EtOOH, H2, H2O2, HACET, HBr, HCHO, HCOOH, HCl, HO2, HO2NO2, HOCl, HONO, HONO2, ISON, ISOOH, MACR, MACR, MACROOH, MGLY, MPAN, Me2CO, Me2CO, MeCHO, MeCO2H, MeCO3H, MeCOCH2OOH, MeCOCH2OOH, MeOH, MeONO2, MeOOH, MeOOH, NALD, NO3, O3, OClO, OH, PAN, PPAN, i-PrOOH, i-PrOOH, n-PrOOH, n-PrOOH, NO, NO3, NO, NO3, O(3P), OH, OH, NO3, OH, O(3P), OH, O(3P), OH, O3, H2O, OH, O3, NO3, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, RO2, EtCO3, O2, NO, NO2, N2, NO2, ClO, NO2, O2, HO2, NO2, NO, NO2, OH, NO2, NO2, NO2, NO3, NO, OH, OH, OH, H2O, HCl, HCl, H2O, HCl, H2O2, O3, O3 # REACT 1, ClO, NO2, MeCO3, EtCO3, MACRO2, NO2, MeCHO, OH, HO2, H2, HO2, OH, MeOO, CH4, HO2, NO3, NO, NO, NO2, O(3P), O2, O2, MeCO3, OH, EtOO, MeCO3, EtCHO, Me2CO, MeCO3, EtCO3, HO2, OH, NO2, MeCO3, MACRO2, OH, HACET, MeCO3, MeCO3, HCHO, MeOO, Br, Br, Br, BrO, O(3P), O(3P), N, OH, N2, OH, Cl, ClO, H, OH, Cl, Cl, Cl, Br, MeOO, CO, OH, COS, CO, SO3, SO2, BrCl, HBr, HBr, BrO, BrO, Br, Br, Br, BrCl, HOBr, Br, Br, Cl, Cl, HCl, Cl, Cl, HCl, HCl, HCl, ClO, HCl, Cl, HCl, ClO, ClO, ClO, Cl, Cl, Cl, HOCl, Cl, Cl, Cl, EtOO, EtOO, MeCHO, MeCHO, H2, O(3P), OH, OH, OH, O2, O3, EtOOH, H2O2, ISOOH, MACROOH, MeCO2H, MeCO3H, OH, MeCOCH2OOH, HCHO, MeOOH, OH, OH, OH, i-PrOOH, n-PrOOH, Br, Br, Br, MeOO, MeOO, MeCO3, MeCO3, HO2, MeONO2, HO2, N2, N2O, NO, HONO2, ISON, NO2, MGLY, NO2, NO2, NO2, NO, NO3, BrO, ISON, HONO2, HONO2, MeCO3, HONO2, HONO2, HCHO, HCHO, OH, O(3P), OH, OH, HBr, OH, H, O(3P), OH, O(3P), N2, NO, O(3P), O2, O2, O2, Cl, ClO, OH, OH, OH, OH, OH, NO, O2, O2, O2, O2, HO2, MACR, MeOO, MGLY, MGLY, OH, OH, NO2, H2O, i-PrOO, n-PrOO, ISO2, H2O, H, HCl, HO2, HOCl, H2O, H2O, H2O, H2O, H2O, MGLY, H2O, H2O, CO2, H2O, H2O, H2O, ClO, H2O, H2O, HACET, MACR, MACRO2, MACRO2, MACRO2, MeCO3, HACET, H2O, H2O, H2O, MeOO, MeCO3, H2O, OH, HO2, HCHO, H2O, H2O, HCHO, HO2, HO2, HOCl, H2O, HCHO, MeCHO, Me2CO, i-PrOO, EtCHO, n-PrOO, Me2CO, Me2CO, EtCHO, EtCHO, CO, COS, SO2, SO2, SO2, OH, SO2, CO, CO2, SO3, H2SO4, Sec_Org, Sec_Org, Sec_Org, HO2, MeOH, HO2, MeCHO, HO2, EtCHO, HO2, Me2CO, MeCO2H, , HCHO, HACET, MACR, HACET, HO2, OH, O3, NO2, NO3, N2O, BrONO2, Cl2O2, ClONO2, HO2, H2O2, HO2NO2, HONO, HONO2, H2O2, PAN, PPAN, MPAN, N2O5, NO2, SO3, DMSO, SO2, HOCl, Cl, Cl, HONO2, Cl, , , # REACT 2, ClO, HO2, NO2, NO2, NO2, NO3, HO2, OH, HO2, CO, NO2, NO2, HO2, CO, HCHO, NO2, O(3P), O2, O(3P), O(3P), O(1D), O(3P), NO2, NO, HO2, MeOO, HO2, HO2, HCHO, NO2, HCHO, MACR, MACR, HCHO, NO2, HO2, CO, HCHO, CO, CO, OH, Cl, O(3P), NO3, NO2, O(1D), ClO, O(3P), Br, O(1D), H, NO3, NO2, Cl, Cl, Cl, Cl, Cl, H, H, O(3P), NO3, SO2, SO2, OH, O(3P), Cl, CO, O2, O2, ClO, Br, Cl, OClO, O2, O2, NO2, HO2, ClO, Cl, MeOO, Cl, Cl, H, HO2, CO, OH, O2, Cl, MeOO, NO2, O2, ClO, Cl, Cl, OClO, O2, HCHO, NO2, O2, CO2, CO2, HO2, HO2, O2, H2O, OH, NO, O2, , , , , , , O3, , MeOO, , , , NO2, NO2, O2, , , HCl, OH, H2O, CO2, CO2, HCHO, HCHO, HCHO, , HCHO, O(3P), O(3P), O(3P), HONO2, , MACR, HCHO, MeCO3, NO2, , NO2, , NO2, , EtCO3, HO2, CO, MeCOCH2OO, MeCO3, H2, HO2, MeOO, CO2, H, OH, O(3P), Br, ClO, HCl, Cl, N2, O2, NO, O2, O(3P), O2, Br, O2, NO3, HO2, Br, Cl, O2, ClO, O2, NO2, O2, ClO, H, OH, HCHO, HCOOH, HCOOH, HCOOH, MeCO3, MeCO3, ClO, EtOO, H2O, H2O, , MeOO, CO2, O2, Cl, NO3, EtCO3, EtOO, MeCHO, H, HO2, HO2, Br, HO2, H2O, Cl, , NO2, H2O, NO2, NO3, NALD, OH, , , , CO, NO2, MeCOCH2OO, MeCOCH2OO, MeCO3, , , MeCOCH2OO, MGLY, HCHO, NO2, HCHO, MeOO, CO, NO2, O2, O2, O(3P), NO2, NO2, OH, H2O, H2O, H2O, HO2, HO2, HO2, HO2, SO2, SO2, , , MSA, SO2, H2O, SO2, SO2, , H2O, , , , HCHO, HCHO, MeCHO, , EtCHO, , Me2CO, , MeOO, EtOO, MeCO3, MGLY, HCHO, MGLY, , EtOO, , , , , , , , , O2, , , , , , , , , NO2, HO2, HO2, MeOO, HONO2, Cl, Cl, HONO2, NO2, , , # REACT 3, , , , , , , OH, , CO, , , , CO, , OH, , , , , , , , , , CO, , OH, OH, OH, , NO2, HCHO, HCHO, CO, , OH, MGLY, HO2, HO2, NO2, , , , , , , , , , , , , , , , O2, Cl, , , , , , , , H, , O2, HO2, , , , O2, O2, , , , , , , ClO, , Cl, NO3, , , HO2, , , OH, , , , , O2, O2, , , HO2, , NO2, NO2, NO2, NO2, NO2, , , , , , , , , , , , , , , , , , , , , , , , , , NO2, NO2, NO2, NO2, NO2, , NO2, , , , , , HCHO, HO2, HACET, , , O2, , , , , CO, HONO2, , , , HO2, , , , , , , , , , , , , , O(3P), , , , , , , , , , , , , , , , MACRO2, CO, HO2, HO2, , , , , , , , , , , , , , , OH, , , , , CO, H, , , O2, , , , , , , , , , , , , , , , , , , H2O, OH, , NO2, , , , , H2O, H2O, , , OH, , NO2, NO2, NO2, NO2, SO2, , , , , OH, , , , , , , , , , , , , , , , , , , , , HO2, HCHO, , CO2, , , , , , , , , , , , , , , , , , , , , , , HONO2, H2O, , HONO2, , , # REACT 4, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , HO2, HO2, HO2, , HO2, HCHO, , , HO2, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , HO2, , CO, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , MeCO3, H2O2, CO, CO, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , O(3P), , , , , , , , , , , , , , , , , , , , , , CO, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , ,
Jupyter Notebook
An example Jupyter Notebook which plots both tracers and chemical reaction fluxes can be found in
Tutorials/UMvn13.0/notebooks/BoxModel_T01-03.ipynb
and can be viewed online here
You can try changing the chemical species and reaction flux selected.
Connect to the notebook using
Checklist
- ☐ List suites using rosie go
- ☐ Copy suites using the right-click menu
- ☐ Run suites using the play button
- ☐ Plot output using gnuplot or python
UKCA Chemistry and Aerosol Tutorials at UMvn13.0
Written by Luke Abraham 2022