Box model
UKCA Box Model - Version 1
The UKCA Box Model is a set of wrapper code which allow the running of the UKCA code base, taken directly from the repository trunk, independently of the UM as if running the chemistry and aerosol code over a single grid cell. The box model is designed to be used primarily for development and testing purposes - due to it's simple design it can quickly compiled and run on a single core in minutes, making debugging of changes to the core chemistry routines substantially faster than on the 3D model. The model has been tested to run in three broad modes, currently it only works with chemical mechanisms which utilise the ASAD framework:
- Chemistry only, using StratTrop or CRI-Strat chemical mechanisms
- GLOMAP Aerosol with offline oxidents
- Coupled online chemistry-aerosol, using GLOMAP-MODE aerosol and StratTrop or CRI-Strat chemical mechanisms.
As well as use for development and testing, the box model is an integral part of the broader UKCA development strategy, as part of the ExCALIBUR ['LINK'] project, to enable the wholesale movement of the UKCA code base out of the UM into a seperpate repository. This transfer is occurring as part of the changes being made for UM vn13.0, Ticket #6717.
Overview
The source code for the box model can be found as a branch of the UKCA repository - in time these will merged into a single trunk which contains the UKCA source code and code to run the box model.
The build options are defined in the file fcm_make/inc/ukca-box-common.cfg; UKCA files/directories used for the build are defined under extract.path-incl[ukca] =
Files defined in the directory src/control/box_control are the box model code wrapping the core UKCA code. The UKCA code based is those under the sections src/control/core and src/science. Currently, the box model still has dependencies on the full UM code base (defined by extract.path-incl[um]). These will be removed and replaced with appropriate replications of functionalism not currently called.
The box model is designed to run like a single grid cell in the full UM model, with no calls to transport or physics but a full integration of the UKCA code base on each timestep. Because of its simple setup, the UKCA box-model typically takes a few minutes for a clean compile and seconds to integrate several days on a single core. This makes it an ideal test bed to for developing, debugging, testing and optimising new functionality into the UKCA model before use in a 3D model.
As the UKCA box-model does not does not compute the changing atmospheric, land and radiation conditions that would usually be calculated by the parent model, it instead has a set of environmental parameters predefined by the user to dictate the environmental conditions and latitude-longitude location of the grid cell being integrated. It also reads in initial mass mixing ratio (MMR, units kg/kg) values for all chemical tracers (and aerosol tracers if running with GLOMAP-mode).
Currently (as UMvn13.0) there are no emissions or dry/wet deposition functionality in the box model. Because of this, integration of the box-model simply progresses from the initial conditions towards a steady state. The box-model is also currently unable to use Fast-JX photolysis due to a lack of column ozone and sulfate aerosol values that are usually provided by the parent model. Instead, it uses a simple photolysis scheme is which j-rate values for different photolysis reactions are provided in a file by the user. The UKCA box-model then applies diurnal cycle by multiplying the raw j-rates by a factor equal to the COS of the solar zenith angle. We plan to add the functionality for simple emissions and deposition, as well as the ability to call Fast-JX using external files for column ozone and sulfate data, as this increased capability would make box-model more suitable for use in scientific analysis.
Compiling and Running
Code branches and necessary locations
UMvn12.2 equivalent
The latest UMvn12.2-equivalent working branch can be found at:
This branch is designed to run with the first iteration of the UKCA repository, which can be found:
https://code.metoffice.gov.uk/trac/um/browser/main/branches/dev/lukeabraham/vn12.2_move_ukca
The UMvn12.2 equivalent suites are designed to run using the branch r1_ukcabox_move_UKCA_br_G. You should make a local copy of this branch in a directory called UKCA-Box. This is required because, at the moment, the metadata necessary to define the box model appearance is not committed to the trunk, therefore there needs to be a local copy of r1_ukcabox_move_UKCA_br_G containing the relevant metadata. The suite expects this to be in $HOME/UKCA-Box/r1_ukcabox_move_UKCA_br_G, so it is simplest to download the branch to that directory:
cd ~/ mkdir UKCA-Box cd UKCA-Box fcm checkout fcm:ukca.x_br/dev/scottarchernicholls/r1_ukcabox_move_UKCA_br_G
At this version necessary routines from the UM, JULES, and ShumLib are still compiled into the box model.
At the Met Office, the required compiler options are defined by the branch
https://code.metoffice.gov.uk/trac/ukca/browser/main/branches/dev/lukeabraham/r1_meto_build
as covered in UKCA ticket ukca:#9.
UMvn13.0 equivalent
The latest UMvn13.0-equivalent working branch can be found at:
https://code.metoffice.gov.uk/trac/ukca/browser/main/branches/dev/lukeabraham/um13.0_um_box
The UMvn13.0 equivalent suites are designed to run using the branch um13.0_um_box. You should make a local copy of this branch in a directory called src. This is required because, at the moment, the metadata necessary to define the box model appearance is not committed to the trunk, therefore there needs to be a local copy of um13.0_um_box containing the relevant metadata. The suite expects this to be in $HOME/src/um13.0_um_box, so it is simplest to download the branch to that directory:
cd ~/ mkdir src cd src fcm checkout fcm:ukca.x_br/dev/lukeabraham/um13.0_um_box
At this version necessary routines from the UM, JULES, and ShumLib are currently still compiled into the box model. This should change soon with the only code necessary from the UKCA repository.
Copying, editing, and running suites
The sections below detail suites available for the Met Office Virtual Machine and internal VDI systems.
Decide on which suite you want to use, then make a copy of it using the rosie command, e.g.
rosie copy SUITE-ID
or graphically using rosie go. This new suite can then be opened, edited and run using the rose gui:
cd ~/roses/u-<suite_id> rose edit &
or by double-clicking in the rosie go suite viewer.
Alternatively, the suite options can be edited by modifying the suite.rc and app/ukca/rose-app.conf then running the suite using:
rose suite-run
or by clicking the play button on the right of the menu bar in the Rose GUI. Note that you should always save and close suites prior to editing options with text editors.
Virtual Machine
The UKCA box model was developed to run on the Virtual Machine framework:
Instruction on how to set up the Virtual Machine to run UKCA can be found here:
Once the virtual machine is set up, it should be possible to run the box model.
| Platform | Equivalent UM version | Configuration | Suite-ID | Notes |
|---|---|---|---|---|
| VM | 12.2 | vm-x86-gnu | u-cn637 | Uses metadata settings from $HOME/UKCA-Box/r1_ukcabox_move_UKCA_br_G Uses debug compiler settings |
| VM | 13.0 | vm-x86-gnu | u-cq645 | Uses metadata settings from $HOME/src/um13.0_um_box Uses rigorous compiler settings. Still compiles in the UM, JULES, & ShumLib code. |
Met Office systems
With thanks to Pedro Molina-Jimenez
The UKCA Box Model has also been implemented to run on the Met Office VDI RHEL7 systems. The available suites are listed below. Both meto-x86-gnu and meto-x86-gcc-ifort compiler settings are available. Not all compiler levels have been tested.
| Platform | Equivalent UM version | Configuration | Suite-ID | Notes |
|---|---|---|---|---|
| VDI | 12.2 | meto-x86-gnu | u-co643 | Uses debug compiler settings as on the VM Also runs using safe compiler settings |
| VDI | 12.2 | meto-x86-gcc-ifort | u-co636 | Runs uses safe compiler settings Fails using rigorous compiler settings Runs using debug compiler settings, but only produces a single line of output in the tracer_out.csv file (timestep 0) |
| VDI | 13.0 | meto-x86-gnu | u-cq678 | Uses metadata settings from $HOME/src/um13.0_um_box Uses rigorous compiler settings as on the VM Also runs using debug compiler settings, but not safe. The column-call option must be used to prevent issues allocating memory on the call to asad_cdrive. |
| VDI | 13.0 | meto-x86-gcc-ifort | u-cq679 | Uses metadata settings from $HOME/src/um13.0_um_box Uses safe compiler settings. Also uses column-call as in u-cq678. |
Currently it seems that these two suites give identical output when run with these different compilers and compiler levels. However, this may not be the case for all configurations.
Then make a copy of the suite you wish to use:
rosie copy SUITE-ID
The new suite (initially a copy of those listed above) can then be opened, edited and run using the rose gui:
cd ~/roses/u-<suite_id> rose edit &
Inputs
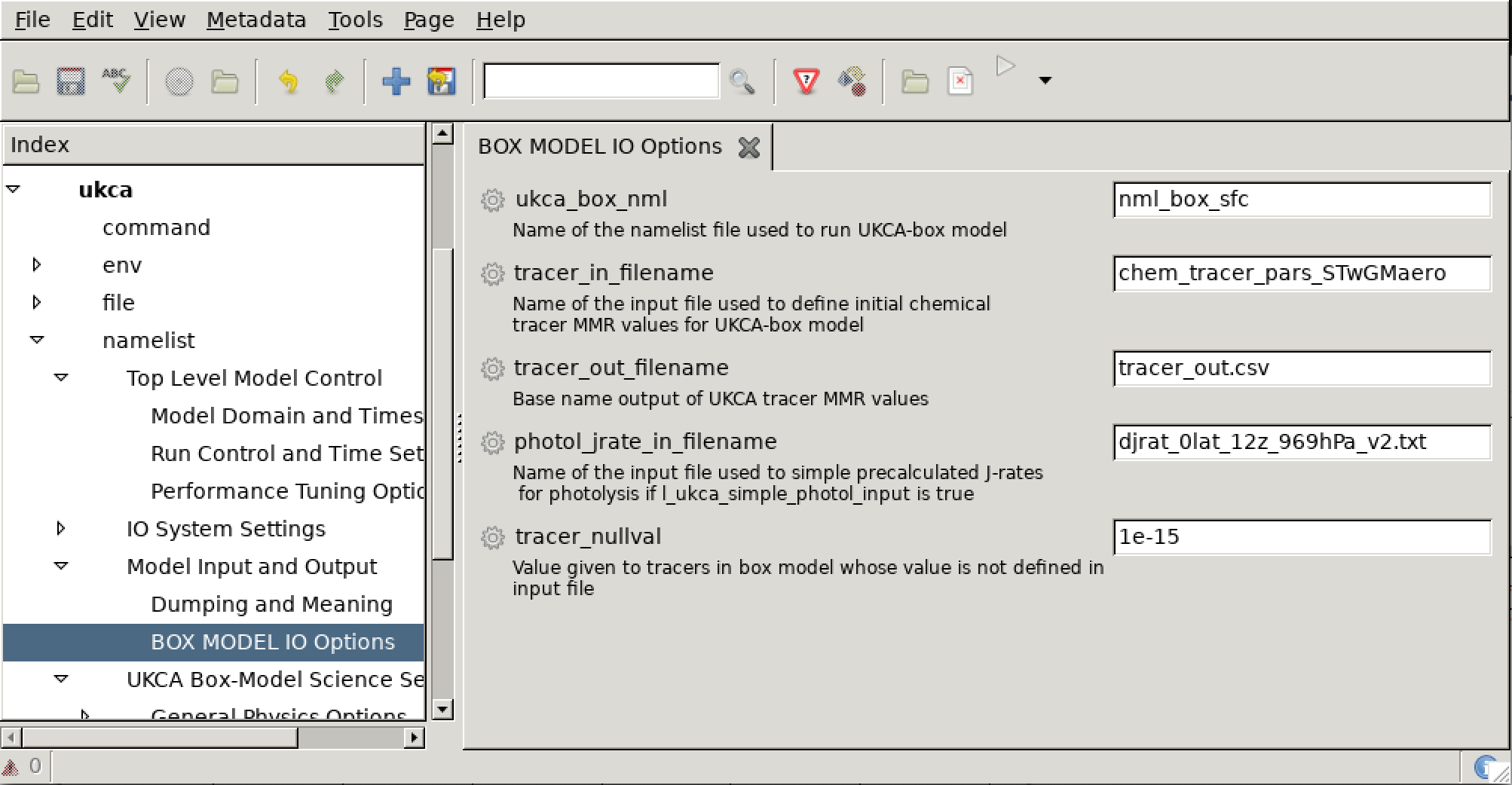
The box model uses a similar setup of rose-meta to the full UM model, with options which no longer apply to the box model (mostly) removed. There is an additional namelist section added to the suite (nlstcall_box_options), which controls inputs required to run the box model and can be found in ukca namelist Model Input and Output BOX MODEL IO Options in Rose. This namelist has the following options:
- ukca_box_nml CHARACTER[*] The name for the external namelist file to define the environmental parameters required to run the UKCA box model
- tracer_in_filename CHARACTER[*] The filename for initial MMR values for chemical and aerosol tracers used by the box model, any undefined tracers will have their initial value set to tracer_nullval
- tracer_out_filename CHARACTER[*] Filename for main output file containing MMR values for each tracer in the box model
- photol_jrate_in_filename CHARACTER[*] Name for input file used by the UKCA box model for precalculated J-Rate values for simple photolysis (only used with StratTrop of CRI-Strat chemistry)
- tracer_nullval REAL The default MMR value for UKCA-box tracers that are not included in the input tracer file (tracer_in_filename)
Most of these options define filenames which contain the initial conditions required to run the box-model. Running a different scenario mostly requires changing one of the above character strings to point to either a different existing file or a new file created by the user. The model will search for all of the files whose filename is defined in the above options in the file directory of the suite being run:
~/roses/<suite_id>/app/ukca/file
The example suite u-cn637 contains the following files in that directory:
- chem_tracer_pars_OffOx Aerosol and aerosol precursor initial condition base case for running with offline oxidents
- chem_tracer_pars_OffOx_LowAero Aerosol and aerosol precursor initial conditions for running with offline oxidents with initial aerosol concentrations reduced by a factor of 10
- chem_tracer_pars_OffOx_NoGP Aerosol initial conditions for running with offline oxidents but with no gasphase precursers.
- chem_tracer_pars_ST_basic Initial tracer MMR values for basic StratTrop setup - only a few species MMRs defined with values typical for clean background troposphere conditions
- chem_tracer_pars_ST_urban Initial MMR values for all gas-phase species in StratTrop typical of clean tropospheric conditions
- chem_tracer_pars_STwGMaero Basic StratTrop initial gas phase concentrations with aerosol and aerosol precurser values the same as Offline Oxident base case
- chem_tracer_pars_STwGMaero_EqTropopause StratTrop initial gas phase concentrations typical of equatorial tropopause with aerosol and aerosol precurser values the same as Offline Oxident base case
- chem_tracer_pars_STwGM_NO3NH4aero_EqTropopause StratTrop initial gas phase concentrations typical of equatorial tropopause with aerosol and aerosol precurser values the same as Offline Oxident base case, including species required for running with ammonium nitrate GLOMAP-mode options
- djrat_0lat_12z_969hPa_v2.txt List of photolysis rates ("j-rates") in units s^-1 for running with simple photolysis, typical for equator near-surface conditions
- nml_box_sfc Namelist options for environmental parameters as if running a grid cell at the surface and 30N
- nml_box_strat Namelist options for environmental parameters as if running a grid cell in the stratosphere at 30N
- nml_box_tropopause Namelist options for environmental parameters as if running a grid cell near the tropopause at 30N
The ukca_box_nml namelist file contains two namelist sections. ENVIRONMENTAL_PARS' must be defined for all setups and defines the location of the box, the pressure (from which is calculated the altitude) and other environmental conditions. OFFLINE_OXIDANT_MMR is only read if running with the offline oxidants mechanism, and defines the MMR values for the oxidant fields. As an example the contents of nml_box_sfc are below:
!----------------------------------------------- ! Box-MODEL test NAMELIST - surface !----------------------------------------------- ! Section to define environmental parameters for box model &ENVIRONMENT_PARS latitude_1 = 30. ! Latitude, degrees North, -90:90 longitude_1 = 0. ! Longitude, degrees East, 0:360 t_theta_levels_1 = 288.15 ! Temperature, K p_theta_levels_1 = 100000. ! Pressure, Pa land_sea_mask_1 = .TRUE. ! Land/sea mask, logical tstar_1 = 288.15 ! Tmperature at surface, K zbl_1 = 1000. ! Height of boundary layer, m pstar_1 = 101325. ! Pressure at surface, Pa q_1 = 0.01 ! Mass fraction of water vapour in air qcf_1 = 0.0 ! Mass fraction of cloud ice in air qcl_1 = 0.0 ! Mass fraction of liquid cloud water in air surf_albedo_1 = 0.2 ! Surface albedo / ! Section to define Offline Oxidant MMR values ! (only used if running with offline oxidants scheme) &OFFLINE_OXIDANT_MMR O3 = 5.0000e-08 OH = 5.0000e-14 HO2 = 5.0000e-12 H2O2 = 5.0000e-10 NO3 = 5.0000e-12 /
In addition to the above namelist options, initial conditions for tracers are provided by one of the chem_tracer_pars files, depending on which chemical mechanism is being used. The file must consist of two columns with a space between them, the first column containing mass mixing ratios (kg/kg) for each species, the second the name of the species to map to. Values can be provided in any order and not all species need to be provided - any not initialised are set to an initial value defined by tracer_nullval (set to 1e-15 by default in u-cn637). The contents of chem_tracer_pars_ST_basic are provided below as an example:
1.0000e-07 O3 5.0000e-10 NO 1.0000e-09 NO2 1.0000e-09 HONO2 1.0000e-09 H2O2 1.0200e-06 CH4 1.0000e-07 CO 1.0000e-12 BrO 1.0000e-12 HCl 1.0000e-11 BrONO2 1.0000e-11 ClONO2 1.0000e-12 BrCl 5.0000e-07 N2O 1.0000e-08 SO2 1.0000e-08 DMS 1.0000e-10 Monoterp
If running with GLOMAP-mode aerosols, initial values for all aerosol tracers must also be provide. These are also in units kg/kg, with the exception of the aerosol number tracers (e.g. Acc_SOL_N) which are in units particles / molecule air. Unlike gas-phase chemical tracers, we recommend explicitly providing initial concentrations for all aerosol species because not all combinations of aerosol mass and number are compatible with each other and the GLOMAP code is unstable if these do not correspond to realistic values. The contents of the chem_tracer_pars_OffOx file are provided below as an example:
1.0000e-09 H2O2 1.0000e-09 DMS 1.0000e-08 SO2 1.0000e-09 Monoterp 5.0000e-17 Nuc_SOL_N 1.0000e-13 Nuc_SOL_SU 5.0000e-17 Ait_SOL_N 5.0000e-10 Ait_SOL_SU 1.0000e-10 Ait_SOL_BC 5.0000e-10 Ait_SOL_OM 1.0000e-17 Acc_SOL_N 5.0000e-09 Acc_SOL_SU 1.0000e-09 Acc_SOL_BC 5.0000e-09 Acc_SOL_OM 1.0000e-10 Acc_SOL_SS 5.0000e-20 Cor_SOL_N 2.0000e-10 Cor_SOL_SU 5.0000e-13 Cor_SOL_BC 5.0000e-12 Cor_SOL_OM 5.0000e-09 Cor_SOL_SS 2.0000e-16 Ait_INS_N 2.0000e-09 Ait_INS_BC 2.0000e-08 Ait_INS_OM 1.0000e-13 Nuc_SOL_OM
Output
Values for the chemical (and aerosol) tracers at each timestep are outputted to the tracer_out_filename file, which can be found in the directory:
~/cylc_run/u-<suite_id>/work/1/ukca/
This is a simple text file of comma separated variables of each of the tracer values in mass mixing ratio units (kg/kg). The top line is a commented out legend naming each of the columns. The first column is timestep (by default one timestep = one hour), each of the following columns are the tracers in the tracer array in the order they appear in the UKCA code.
A simple way to plot these is using Gnuplot. This can be installed on the Virtual Machine using the command:
sudo apt-get install gnuplot
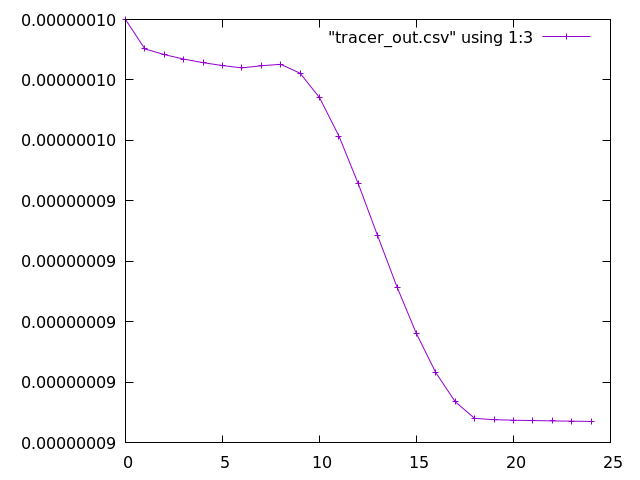
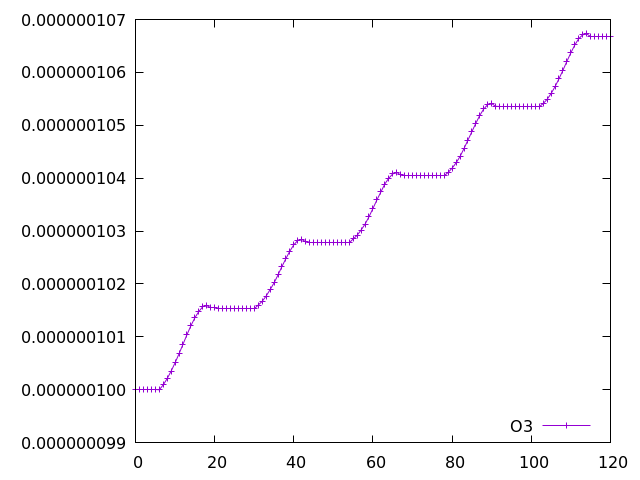
As an example, ozone (the 2nd species in the tracer array, therefore the third column after timestep and O(3P)) can be plotted as follows:
gnuplot set datafile separator "," plot "tracer_out.csv" using 1:3 with linespoints
Additional plotting options for Gnuplot can be supplied, such as using title to assign names to the lines in the legend. Alternative plotting tools, such as Python, can also be used.
Examples
Change the box model run length
Currently the box model uses similar variables to the UM for many of its parameters, including those related to run length and model time. The namelist variable to control the run length of a box model simulation is run_target_end, which can be found in the Run Control and Time Settings panel located at ukca namelist Top Level Model Control Run Control and Time Settings.
It is configured as 6 boxes corresponding to
Years, Months, Days, Hours, Minutes, Seconds
and has an initial value of 0,0,1,0,0,0. You can change this to be, e.g. 0,0,5,0,0,0 to give a run length of 5 days, then save and run your suite.
Change the scenario
As covered in the Inputs section above, the box model scenario is defined by files specified in the ukca namelist Model Input and Output BOX MODEL IO Options panel, as can be seen below
If we change these values to be
tracer_in_filename='chem_tracer_pars_STwGMaero_EqTropopause' ukca_box_nml='nml_box_tropopause'
we will now be running the StratTrop at the equatorial tropopause, rather than at the surface.
Change the chemical mechanism
It is not strictly necessary to change the tracer_in_filename when changing chemical mechanism, as any additional species defined in the input list will be ignored, and any missing species will be assigned an initial value as defined by tracer_nullval.
The chemical mechanism used is set by the i_ukca_chem parameter found in the ukca namelist UKCA Box-model Science Settings UKCA: UK Aerosols and Chemistry panel.
CRI-Strat
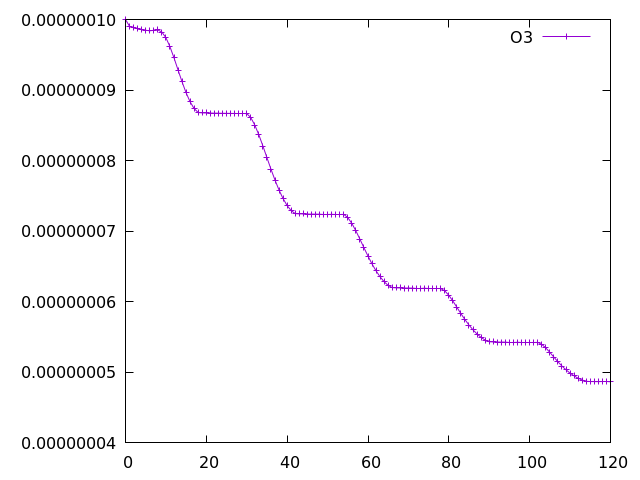
The easiest way to turn on the CRI-Strat scheme is to change the value of i_ukca_chem to 59. Keeping the current tracer_in_filename ('chem_tracer_pars_STwGMaero_EqTropopause') from the above example is absolutely fine as tracer_nullval is set to 1e-15.
The plot of o3 looks very similar to the StratTrop example, but examining the first line of the tracer_out.csv file it can be seen that this contains many more species, corresponding to the CRI-Strat scheme:
#Timestep,O(3P) ,O3 ,N ,NO ,NO3 ,NO2 ,N2O5 ,HO2NO2 ,HONO2 ,H2O2 ,CH4 ,CO ,HCHO ,MeOOH ,H ,H2O ,OH ,HO2 ,Cl ,Cl2O2 ,ClO ,OClO ,Br ,BrO ,BrCl ,BrONO2 ,N2O ,HCl ,HOCl ,HBr ,HOBr ,ClONO2 ,CFCl3 ,CF2Cl2 ,MeBr ,HONO ,C2H6 ,EtOOH ,MeCHO ,PAN ,C3H8 ,i-PrOOH ,EtCHO ,Me2CO ,PPAN ,MeONO2 ,C5H8 ,MPAN ,HCOOH ,MeCO3H ,MeCO2H ,H2 ,MeOH ,DMS ,SO2 ,H2SO4 ,MSA ,DMSO ,NH3 ,CS2 ,COS ,H2S ,SO3 ,Sec_Org ,C2H4 ,C3H6 ,C4H10 ,MEK ,TOLUENE ,oXYLENE ,TBUT2ENE ,APINENE ,BPINENE ,C2H2 ,BENZENE ,EtOH ,i-PrOH ,n-PrOH ,HOCH2CHO ,HOC2H4OOH ,EtCO3H ,HOCH2CO3H ,NOA ,EtONO2 ,i-PrONO2 ,MeO2NO2 ,HOC2H4NO3 ,PHAN ,MeSCH2OO ,MeS ,MeSO ,MeSO2 ,MeSO3 ,MSIA ,CARB14 ,CARB17 ,CARB11A ,CARB7 ,CARB10 ,CARB13 ,CARB16 ,UCARB10 ,CARB3 ,CARB6 ,CARB9 ,CARB12 ,CARB15 ,UCARB12 ,NUCARB12 ,UDCARB8 ,UDCARB11 ,UDCARB14 ,TNCARB26 ,TNCARB10 ,RN10NO3 ,RN13NO3 ,RN16NO3 ,RN19NO3 ,RA13NO3 ,RA16NO3 ,RA19NO3 ,RTX24NO3 ,RN10OOH ,RN13OOH ,RN16OOH ,RN19OOH ,RN8OOH ,RN11OOH ,RN14OOH ,RN17OOH ,RU14OOH ,RU12OOH ,RU10OOH ,NRU14OOH ,NRU12OOH ,RN9OOH ,RN12OOH ,RN15OOH ,RN18OOH ,NRN6OOH ,NRN9OOH ,NRN12OOH ,RA13OOH ,RA16OOH ,RA19OOH ,RTN28OOH ,NRTN28OOH ,RTN26OOH ,RTN25OOH ,RTN24OOH ,RTN23OOH ,RTN14OOH ,RTN10OOH ,RTX28OOH ,RTX24OOH ,RTX22OOH ,NRTX28OOH ,RAROH14 ,RAROH17 ,RU12PAN ,RTN26PAN ,TNCARB12 ,TNCARB11 ,RTN23NO3 ,CCARB12 ,TNCARB15 ,RCOOH25 ,TXCARB24 ,TXCARB22 ,RN9NO3 ,RN12NO3 ,RN15NO3 ,RN18NO3 ,RU14NO3 ,RTN28NO3 ,RTN25NO3 ,RTX28NO3 ,RTX22NO3 ,AROH14 ,ARNOH14 ,AROH17 ,ARNOH17 ,ANHY ,Nuc_SOL_N ,Nuc_SOL_SU,Ait_SOL_N ,Ait_SOL_SU,Ait_SOL_BC,Ait_SOL_OM,Acc_SOL_N ,Acc_SOL_SU,Acc_SOL_BC,Acc_SOL_OM,Acc_SOL_SS,Cor_SOL_N ,Cor_SOL_SU,Cor_SOL_BC,Cor_SOL_OM,Cor_SOL_SS,Ait_INS_N ,Ait_INS_BC,Ait_INS_OM,Nuc_SOL_OM
Offline-oxidants
The offline-oxidants scheme should be selected using i_ukca_chem=54 (Note: NOT 14 here, which corresponds to using a different chemical solver). For this example we will also change the settings in the ukca namelist Model Input and Output BOX MODEL IO Options panel to use ukca_box_nml='nml_box_sfc' to select the surface and tracer_in_filename='chem_tracer_pars_OffOx' to define the initial conditions for the tracers and GLOMAP-mode aerosols. Looking at the top of the tracer_out.csv file we can see the change in scheme:
#Timestep,H2O2 ,DMS ,SO2 ,H2SO4 ,DMSO ,Monoterp ,Sec_Org ,Nuc_SOL_N ,Nuc_SOL_SU,Ait_SOL_N ,Ait_SOL_SU,Ait_SOL_BC,Ait_SOL_OM,Acc_SOL_N ,Acc_SOL_SU,Acc_SOL_BC,Acc_SOL_OM,Acc_SOL_SS,Cor_SOL_N ,Cor_SOL_SU,Cor_SOL_BC,Cor_SOL_OM,Cor_SOL_SS,Ait_INS_N ,Ait_INS_BC,Ait_INS_OM,Nuc_SOL_OM
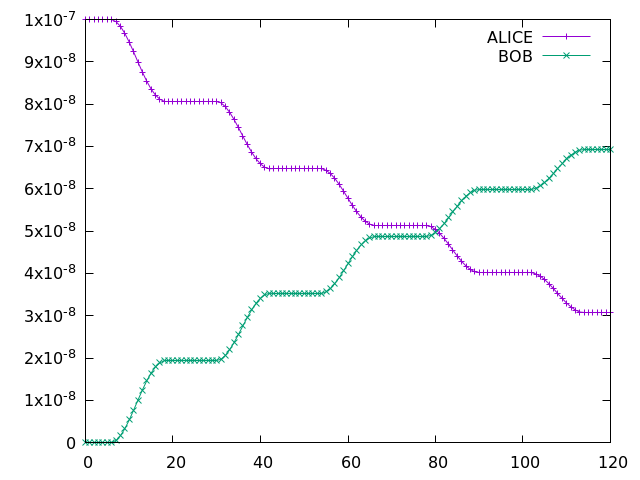
Adding new chemical species - ALICE and BOB
Following the instructions from the UKCA Tutorials it is straight-forward to add new species and reactions. Currently emissions and deposition processes are not included in the Box model, so cannot be included at this stage.
Following the UMvn11.8 UKCA Tutorials, the new species ALICE and BOB can be added, and the reaction
| Parameter | Value |
|---|---|
| 2.70E-11 | |
| 0.00 | |
| -390.00 |
can be included. As there are no emissions, the initial conditions of ALICE can be set by editing the file specified in tracer_in_filename (e.g. 'chem_tracer_pars_STwGMaero') to include the line, e.g.
1.0000e-07 ALICE
Until code is committed to the UKCA trunk, these changes must be made in a copy of the branch fcm:ukca.x_br/dev/lukeabraham/r1_move_ukca. The code changes are below, covering adding the ALICE and BOB tracers and the reaction between them:
Index: src/control/core/top_level/ukca_config_defs_mod.F90
===================================================================
--- src/control/core/top_level/ukca_config_defs_mod.F90 (revision 243)
+++ src/control/core/top_level/ukca_config_defs_mod.F90 (revision 245)
@@ -315,7 +315,7 @@
['NO ','CH4 ','CO ','HCHO ', &
'C2H6 ','C3H8 ','Me2CO ','MeCHO ', &
'C5H8 ', nm_meoh_em ,'NO_aircrft']
- n_chem_tracers = 71 ! No chem tracers
+ n_chem_tracers = 73 ! No chem tracers
nr_therm = 220 ! thermal reactions
nr_phot = 55 ! photolytic (ATA)
@@ -331,11 +331,11 @@
'SO2_high ','NH3 ','DMS ','SO2_nat ', &
'BC_biomass','OM_biomass','NO_aircrft']
n_aero_tracers = 12
- n_chem_tracers = 71 ! No chem tracers
+ n_chem_tracers = 73 ! No chem tracers
IF (ukca_config%l_ukca_trophet) THEN
- nr_therm = 241 ! thermal reactions
+ nr_therm = 242 ! thermal reactions
ELSE
- nr_therm = 239 ! thermal reactions
+ nr_therm = 240 ! thermal reactions
END IF
nr_phot = 59 ! photolytic (ATA)
Index: src/science/core/chemistry/ukca_chem_master.F90
===================================================================
--- src/science/core/chemistry/ukca_chem_master.F90 (revision 243)
+++ src/science/core/chemistry/ukca_chem_master.F90 (revision 245)
@@ -103,11 +103,11 @@
! later ones
! Define size of master chemistry
-INTEGER, PARAMETER :: n_chch_master = 356 ! number of known species
+INTEGER, PARAMETER :: n_chch_master = 358 ! number of known species
INTEGER, PARAMETER :: n_het_master = 18 ! number of heterogeneous reactions
INTEGER, PARAMETER :: n_dry_master = 161 ! number of dry deposition reactions
INTEGER, PARAMETER :: n_wet_master = 159 ! number of wet deposition reactions
-INTEGER, PARAMETER :: n_bimol_master = 1209 ! number of bimolecular reactions
+INTEGER, PARAMETER :: n_bimol_master = 1210 ! number of bimolecular reactions
INTEGER, PARAMETER :: n_ratj_master = 183 ! number of photolysis reactions
INTEGER, PARAMETER :: n_ratt_master = 116 ! number of termolecular reactions
@@ -917,7 +917,11 @@
! 282
chch_t1(282,'RU10NO3 ',1,'TR ',' ',1,1,cs,0,0,119), &
! 283
-chch_t1(283,'MACO3 ',1,'OO ',' ',0,0,cs,rp,0,119) &
+chch_t1(283,'MACO3 ',1,'OO ',' ',0,0,cs,rp,0,119), &
+! 284
+chch_t1(284,'ALICE ',1,'TR ',' ',0,0,st,0,0,130), &
+! 285
+chch_t1(285,'BOB ',1,'TR ',' ',0,0,st,0,0,130) &
]
@@ -6223,7 +6227,10 @@
' ',3.93e-15, 0.00, 1913.00, 0.54, 0.00, 0.00, 0.00,r,0,0,107), &
! B745 - updates for 121
ratb_t1(745,'HO2 ','EtCO3 ','OH ','EtOO ','CO2 ', &
-' ',1.89e-13, 0.00, -1040.00, 0.00, 0.00, 0.00, 0.00,st,0,0,121) &
+' ',1.89e-13, 0.00, -1040.00, 0.00, 0.00, 0.00, 0.00,st,0,0,121), &
+! B746 - updates for 130 (ALICE & BOB tutorials)
+ratb_t1(746,'ALICE ','OH ','BOB ','Sec_Org ',' ', &
+' ',2.70e-11, 0.00, -390.00, 0.00, 0.00, 0.00, 0.00,st,a,0,130) &
]
!----------------------------------------------------------------------
Index: src/science/core/chemistry/ukca_constants.F90
===================================================================
--- src/science/core/chemistry/ukca_constants.F90 (revision 243)
+++ src/science/core/chemistry/ukca_constants.F90 (revision 245)
@@ -357,6 +357,11 @@
! Same as Sec_org
REAL, PARAMETER :: c_cri = 5.1782
+! define conversion factors for tutorial species
+REAL, PARAMETER :: c_alice = 1.000
+REAL, PARAMETER :: c_bob = 1.000
+
+
! molecular masses in g/mol of emitted species,
! for budget calculations
REAL, PARAMETER :: m_ho2 = 33.007
Index: src/science/core/chemistry/ukca_cspecies.F90
===================================================================
--- src/science/core/chemistry/ukca_cspecies.F90 (revision 243)
+++ src/science/core/chemistry/ukca_cspecies.F90 (revision 245)
@@ -182,7 +182,7 @@
c_txcarb22, c_ru12pan, c_rtn26pan, c_aroh14, c_aroh17, c_arnoh14, &
c_arnoh17, c_anhy, c_cri,c_dhpcarb9, c_hpucarb12, c_hucarb9, c_iepox, &
c_hmml, c_dhpr12ooh, c_dhcarb9, c_ru12no3, c_ru10no3, c_dhpr12o2, &
- c_ru10ao2, c_maco3
+ c_ru10ao2, c_maco3, c_alice, c_bob
USE asad_mod, ONLY: advt, nadvt, speci, jpctr, jpspec
@@ -455,6 +455,9 @@
WHERE (advt == 'DHCARB9 ') c_species = c_dhcarb9
WHERE (advt == 'RU12NO3 ') c_species = c_ru12no3
WHERE (advt == 'RU10NO3 ') c_species = c_ru10no3
+! tutorial species
+WHERE (advt == 'ALICE ') c_species = c_alice
+WHERE (advt == 'BOB ') c_species = c_bob
! Mass for these species does not matter as not emitted or deposited
! Use default mass 150g/mole
In the Rose suite, the value of i_ukca_chem_version should be set to 130 for these changes to be picked-up.
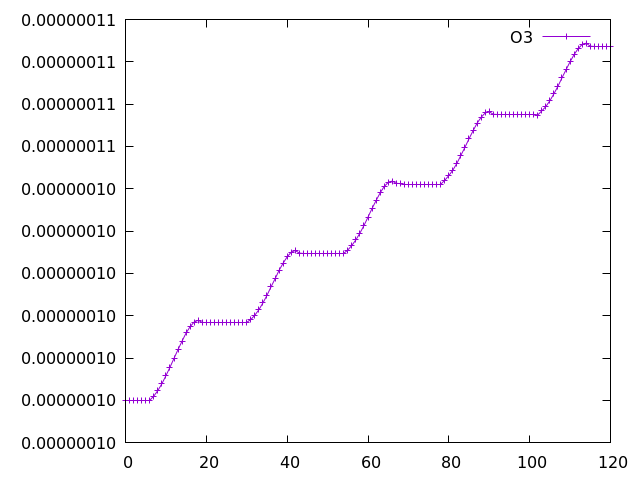
Looking in the tracer_out.csv file we can see that ALICE and BOB now exist as output:
#Timestep,O(3P) ,O3 ,N ,NO ,NO3 ,NO2 ,N2O5 ,HO2NO2 ,HONO2 ,H2O2 ,CH4 ,CO ,HCHO ,MeOOH ,H ,H2O ,OH ,HO2 ,Cl ,Cl2O2 ,ClO ,OClO ,Br ,BrO ,BrCl ,BrONO2 ,N2O ,HCl ,HOCl ,HBr ,HOBr ,ClONO2 ,CFCl3 ,CF2Cl2 ,MeBr ,HONO ,C2H6 ,EtOOH ,MeCHO ,PAN ,C3H8 ,n-PrOOH ,i-PrOOH ,EtCHO ,Me2CO ,MeCOCH2OOH,PPAN ,MeONO2 ,C5H8 ,ISOOH ,ISON ,MACR ,MACROOH ,MPAN ,HACET ,MGLY ,NALD ,HCOOH ,MeCO3H ,MeCO2H ,H2 ,MeOH ,DMS ,SO2 ,H2SO4 ,MSA ,DMSO ,NH3 ,CS2 ,COS ,H2S ,SO3 ,Monoterp ,Sec_Org ,ALICE ,BOB ,Nuc_SOL_N ,Nuc_SOL_SU,Ait_SOL_N ,Ait_SOL_SU,Ait_SOL_BC,Ait_SOL_OM,Acc_SOL_N ,Acc_SOL_SU,Acc_SOL_BC,Acc_SOL_OM,Acc_SOL_SS,Cor_SOL_N ,Cor_SOL_SU,Cor_SOL_BC,Cor_SOL_OM,Cor_SOL_SS,Ait_INS_N ,Ait_INS_BC,Ait_INS_OM,Nuc_SOL_OM,PASSIVE O3
with the species at columns 76 (ALICE) and 77 (BOB). Running a 5-day simulation has the following output:
Example Gnuplot plot of ALICE and BOB from a 5-day simulation implementing the UMvn11.8 UKCA Tutorials into the box model.
Outstanding Tasks
These are outstanding tasks and additional functionality that would be good to add to the box model:
- Add more standard initial condition files for different chemical mechanisms, environmental conditions and photolysis conditions
- Potentially split initial chemical and aerosol tracer initial condition files
- Once UMvn13.0 is released, update so that it can be compiled without using the UM trunk
- Update metadata and refresh the suite accordingly
- Add further examples
- More output options analysing output e.g. fluxes through reactions, production/loss of species
- New scientific functionality:
- Dry deposition
- Wet deposition
- Emissions
- Photolysis using Fast-JX
- Boundary layer dynamics
- Rose suite changes to allow for ensembles of box models, potentially with random initial condition variation for Monte-Carlo methods
- Debug the Ifort errors seen on the Met Office VDIs. This seems to be associated with a call to exitchek (currently a UM module) from the box_model.F90 routine